Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells
- PMID: 22387999
- PMCID: PMC3538378
- DOI: 10.1038/ng.1110
Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells
Abstract
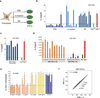
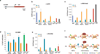
The generation of induced pluripotent stem cells (iPSCs) often results in aberrant epigenetic silencing of the imprinted Dlk1-Dio3 gene cluster, compromising the ability to generate entirely iPSC-derived adult mice ('all-iPSC mice'). Here, we show that reprogramming in the presence of ascorbic acid attenuates hypermethylation of Dlk1-Dio3 by enabling a chromatin configuration that interferes with binding of the de novo DNA methyltransferase Dnmt3a. This approach allowed us to generate all-iPSC mice from mature B cells, which have until now failed to support the development of exclusively iPSC-derived postnatal animals. Our data show that transcription factor-mediated reprogramming can endow a defined, terminally differentiated cell type with a developmental potential equivalent to that of embryonic stem cells. More generally, these findings indicate that culture conditions during cellular reprogramming can strongly influence the epigenetic and biological properties of the resultant iPSCs.
Figures



Comment in
-
Vitamin C improves the quality of somatic cell reprogramming.Nat Genet. 2012 Mar 28;44(4):366-7. doi: 10.1038/ng.2222. Nat Genet. 2012. PMID: 22456737
References
-
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. - PubMed
-
- da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–316. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

