Importance of Micromonospora spp. as colonizers of cellulose in freshwater lakes as demonstrated by quantitative reverse transcriptase PCR of 16S rRNA
- PMID: 22389367
- PMCID: PMC3346464
- DOI: 10.1128/AEM.07314-11
Importance of Micromonospora spp. as colonizers of cellulose in freshwater lakes as demonstrated by quantitative reverse transcriptase PCR of 16S rRNA
Abstract
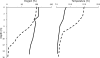
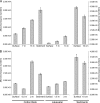
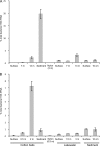
The relative abundance of micromonosporas in the bacterial communities inhabiting cellulose baits, water columns, and sediments of two freshwater lakes was determined by quantitative PCR (qPCR) of reverse-transcribed 16S rRNA. Micromonospora spp. were shown to be significant members of the active bacterial population colonizing cellulosic substrates in the lake sediment, and their increased prevalence with greater depth was confirmed by enumeration of CFU.
Figures
References
-
- Cross T. 1981. Aquatic actinomycetes—a critical survey of the occurrence, growth and role of actinomycetes in aquatic habitats. J. Appl. Bacteriol. 50:397–423 - PubMed
-
- Finlay BJ, Maberly SC. 2000. Microbial diversity in Priest Pot: a productive pond in the English Lake District. Freshwater Biological Association special publication 9. Freshwater Biological Association, Ambleside, Cumbria, United Kingdom
-
- George DG, Talling JF, Rigg E. 2000. Factors influencing the temporal coherence of five lakes in the English Lake District. Freshwater Biol. 43:449–461
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

