Direct binding of a hepatitis C virus inhibitor to the viral capsid protein
- PMID: 22389688
- PMCID: PMC3289641
- DOI: 10.1371/journal.pone.0032207
Direct binding of a hepatitis C virus inhibitor to the viral capsid protein
Abstract
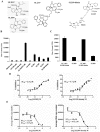
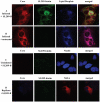
Over 130 million people are infected chronically with hepatitis C virus (HCV), which, together with HBV, is the leading cause of liver disease. Novel small molecule inhibitors of Hepatitis C virus (HCV) are needed to complement or replace current treatments based on pegylated interferon and ribavirin, which are only partially successful and plagued with side-effects. Assembly of the virion is initiated by the oligomerization of core, the capsid protein, followed by the interaction with NS5A and other HCV proteins. By screening for inhibitors of core dimerization, we previously discovered peptides and drug-like compounds that disrupt interactions between core and other HCV proteins, NS3 and NS5A, and block HCV production. Here we report that a biotinylated derivative of SL209, a prototype small molecule inhibitor of core dimerization (IC(50) of 2.80 µM) that inhibits HCV production with an EC(50) of 3.20 µM, is capable of penetrating HCV-infected cells and tracking with core. Interaction between the inhibitors, core and other viral proteins was demonstrated by SL209-mediated affinity-isolation of HCV proteins from lysates of infected cells, or of the corresponding recombinant HCV proteins. SL209-like inhibitors of HCV core may form the basis of novel treatments of Hepatitis C in combination with other target-specific HCV drugs such as inhibitors of the NS3 protease, the NS5B polymerase, or the NS5A regulatory protein. More generally, our work supports the hypothesis that inhibitors of viral capsid formation might constitute a new class of potent antiviral agents, as was recently also shown for HIV capsid inhibitors.
Conflict of interest statement
Figures
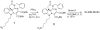

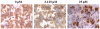
Similar articles
-
Core as a novel viral target for hepatitis C drugs.Viruses. 2010 Aug;2(8):1734-1751. doi: 10.3390/v2081734. Epub 2010 Aug 20. Viruses. 2010. PMID: 21994704 Free PMC article.
-
[Determination of the in vitro antiviral activity of an engineered M1GS ribozyme that targets to the core gene of hepatitis C virus].Wei Sheng Wu Xue Bao. 2013 Aug 4;53(8):875-81. Wei Sheng Wu Xue Bao. 2013. PMID: 24341280 Chinese.
-
In silico studies of medicinal compounds against hepatitis C capsid protein from north India.Bioinform Biol Insights. 2014 Jun 23;8:159-68. doi: 10.4137/BBI.S15211. eCollection 2014. Bioinform Biol Insights. 2014. PMID: 25002815 Free PMC article.
-
Hepatitis C virus resistance to protease inhibitors.J Hepatol. 2011 Jul;55(1):192-206. doi: 10.1016/j.jhep.2011.01.011. Epub 2011 Feb 1. J Hepatol. 2011. PMID: 21284949 Review.
-
Discovery and development of VX-950, a novel, covalent, and reversible inhibitor of hepatitis C virus NS3.4A serine protease.Infect Disord Drug Targets. 2006 Mar;6(1):3-16. doi: 10.2174/187152606776056706. Infect Disord Drug Targets. 2006. PMID: 16787300 Review.
Cited by
-
Human Transbodies to HCV NS3/4A Protease Inhibit Viral Replication and Restore Host Innate Immunity.Front Immunol. 2016 Aug 26;7:318. doi: 10.3389/fimmu.2016.00318. eCollection 2016. Front Immunol. 2016. PMID: 27617013 Free PMC article.
-
Targeting the Virus Capsid as a Tool to Fight RNA Viruses.Viruses. 2022 Jan 18;14(2):174. doi: 10.3390/v14020174. Viruses. 2022. PMID: 35215767 Free PMC article. Review.
-
HCV core protein and virus assembly: what we know without structures.Immunol Res. 2014 Oct;60(1):1-10. doi: 10.1007/s12026-014-8494-3. Immunol Res. 2014. PMID: 24557493 Free PMC article. Review.
-
Discovery and characterization of a novel HCV inhibitor targeting the late stage of HCV life cycle.Antivir Ther. 2019;24(5):371-381. doi: 10.3851/IMP3303. Antivir Ther. 2019. PMID: 30880685 Free PMC article.
-
The A-Z of Zika drug discovery.Drug Discov Today. 2018 Nov;23(11):1833-1847. doi: 10.1016/j.drudis.2018.06.014. Epub 2018 Jun 20. Drug Discov Today. 2018. PMID: 29935345 Free PMC article. Review.
References
-
- Lavanchy D. Evolving epidemiology of hepatitis C virus. Clinical Microbiol Infect. 2011;17:107–115. - PubMed
-
- Cristina J, Moreno-del Pilar M, Moratorio G. Hepatitis C virus genetic variability in patients undergoing antiviral therapy. Virus Research. 2007;127:185–194. - PubMed
-
- Gentile I, Carleo MA, Borgia F, Castaldo G, Borgia G. The efficacy and safety of telaprevir - a new protease inhibitor against hepatitis C virus. Expert Opi Invest Drugs. 2010;19:151–159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

