Development and characterization of a reverse genetic system for studying dengue virus serotype 3 strain variation and neutralization
- PMID: 22389731
- PMCID: PMC3289595
- DOI: 10.1371/journal.pntd.0001486
Development and characterization of a reverse genetic system for studying dengue virus serotype 3 strain variation and neutralization
Abstract
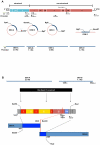
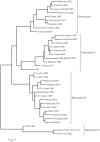
Dengue viruses (DENV) are enveloped single-stranded positive-sense RNA viruses transmitted by Aedes spp. mosquitoes. There are four genetically distinct serotypes designated DENV-1 through DENV-4, each further subdivided into distinct genotypes. The dengue scientific community has long contended that infection with one serotype confers lifelong protection against subsequent infection with the same serotype, irrespective of virus genotype. However this hypothesis is under increased scrutiny and the role of DENV genotypic variation in protection from repeated infection is less certain. As dengue vaccine trials move increasingly into field-testing, there is an urgent need to develop tools to better define the role of genotypic variation in DENV infection and immunity. To better understand genotypic variation in DENV-3 neutralization and protection, we designed and constructed a panel of isogenic, recombinant DENV-3 infectious clones, each expressing an envelope glycoprotein from a different DENV-3 genotype; Philippines 1982 (genotype I), Thailand 1995 (genotype II), Sri Lanka 1989 and Cuba 2002 (genotype III) and Puerto Rico 1977 (genotype IV). We used the panel to explore how natural envelope variation influences DENV-polyclonal serum interactions. When the recombinant viruses were tested in neutralization assays using immune sera from primary DENV infections, neutralization titers varied by as much as ∼19-fold, depending on the expressed envelope glycoprotein. The observed variability in neutralization titers suggests that relatively few residue changes in the E glycoprotein may have significant effects on DENV specific humoral immunity and influence antibody mediated protection or disease enhancement in the setting of both natural infection and vaccination. These genotypic differences are also likely to be important in temporal and spatial microevolution of DENV-3 in the background of heterotypic neutralization. The recombinant and synthetic tools described here are valuable for testing hypotheses on genetic determinants of DENV-3 immunopathogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Neutralizing and enhancing antibody responses to five genotypes of dengue virus type 1 (DENV-1) in DENV-1 patients.J Gen Virol. 2017 Feb;98(2):166-172. doi: 10.1099/jgv.0.000669. Epub 2017 Feb 24. J Gen Virol. 2017. PMID: 27911254
-
Genotype-specific and cross-reactive neutralizing antibodies induced by dengue virus infection: detection of antibodies with different levels of neutralizing activities against homologous and heterologous genotypes of dengue virus type 2 in common marmosets (Callithrix jacchus).Virol J. 2018 Mar 27;15(1):51. doi: 10.1186/s12985-018-0967-x. Virol J. 2018. PMID: 29587780 Free PMC article.
-
Dengue Virus Serotype 1 Conformational Dynamics Confers Virus Strain-Dependent Patterns of Neutralization by Polyclonal Sera.J Virol. 2021 Nov 9;95(23):e0095621. doi: 10.1128/JVI.00956-21. Epub 2021 Sep 22. J Virol. 2021. PMID: 34549976 Free PMC article.
-
A recombinant, chimeric tetravalent dengue vaccine candidate based on a dengue virus serotype 2 backbone.Expert Rev Vaccines. 2016;15(4):497-508. doi: 10.1586/14760584.2016.1128328. Epub 2016 Feb 22. Expert Rev Vaccines. 2016. PMID: 26635182 Review.
-
Dengue Vaccines: The Promise and Pitfalls of Antibody-Mediated Protection.Cell Host Microbe. 2021 Jan 13;29(1):13-22. doi: 10.1016/j.chom.2020.12.011. Cell Host Microbe. 2021. PMID: 33444553 Review.
Cited by
-
Neutralizing antibody correlates of sequence specific dengue disease in a tetravalent dengue vaccine efficacy trial in Asia.Vaccine. 2022 Sep 29;40(41):5912-5923. doi: 10.1016/j.vaccine.2022.08.055. Epub 2022 Sep 5. Vaccine. 2022. PMID: 36068106 Free PMC article. Clinical Trial.
-
DNA vaccine initiates replication of live attenuated chikungunya virus in vitro and elicits protective immune response in mice.J Infect Dis. 2014 Jun 15;209(12):1882-90. doi: 10.1093/infdis/jiu114. Epub 2014 Feb 28. J Infect Dis. 2014. PMID: 24585894 Free PMC article.
-
Construction and characterisation of a complete reverse genetics system of dengue virus type 3.Mem Inst Oswaldo Cruz. 2013 Dec;108(8):983-91. doi: 10.1590/0074-0276130298. Mem Inst Oswaldo Cruz. 2013. PMID: 24402142 Free PMC article.
-
A tetravalent live attenuated dengue virus vaccine stimulates balanced immunity to multiple serotypes in humans.Nat Commun. 2021 Feb 17;12(1):1102. doi: 10.1038/s41467-021-21384-0. Nat Commun. 2021. PMID: 33597521 Free PMC article.
-
The use of longitudinal cohorts for studies of dengue viral pathogenesis and protection.Curr Opin Virol. 2018 Apr;29:51-61. doi: 10.1016/j.coviro.2018.03.004. Epub 2018 Mar 26. Curr Opin Virol. 2018. PMID: 29597086 Free PMC article. Review.
References
-
- Kyle JL, Harris E. Global spread and persistence of dengue. Annu Rev Microbiol. 2008;62:71–92. - PubMed
-
- Burke DS, Nisalak A, Johnson DE, Scott RM. A prospective study of dengue infections in bangkok. Am J Trop Med Hyg. 1988;38:172–180. - PubMed
-
- [Anonymous] Dengue and dengue hemorrhagic fever. Wallingford, Oxon, UK; New York, , NY, USA: CAB International; 1997. Available: http://search.lib.unc.edu?R=UNCb3110999.
-
- Halstead SB. Antibodies determine virulence in dengue. Ann N Y Acad Sci. 2009;1171(Suppl 1):E48–56. - PubMed
-
- Rothman AL. Cellular immunology of sequential dengue virus infection and its role in disease pathogenesis. Curr Top Microbiol Immunol. 2010;338:83–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

