Biochemical, mutational and in silico structural evidence for a functional dimeric form of the ornithine decarboxylase from Entamoeba histolytica
- PMID: 22389745
- PMCID: PMC3289617
- DOI: 10.1371/journal.pntd.0001559
Biochemical, mutational and in silico structural evidence for a functional dimeric form of the ornithine decarboxylase from Entamoeba histolytica
Abstract
Background: Entamoeba histolytica is responsible for causing amoebiasis. Polyamine biosynthesis pathway enzymes are potential drug targets in parasitic protozoan diseases. The first and rate-limiting step of this pathway is catalyzed by ornithine decarboxylase (ODC). ODC enzyme functions as an obligate dimer. However, partially purified ODC from E. histolytica (EhODC) is reported to exist in a pentameric state.
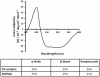
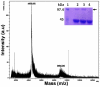
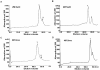
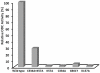
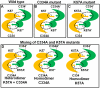
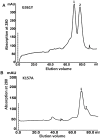
Methodology and results: In present study, the oligomeric state of EhODC was re-investigated. The enzyme was over-expressed in Escherichia coli and purified. Pure protein was used for determination of secondary structure content using circular dichroism spectroscopy. The percentages of α-helix, β-sheets and random coils in EhODC were estimated to be 39%, 25% and 36% respectively. Size-exclusion chromatography and mass spectrophotometry analysis revealed that EhODC enzyme exists in dimeric form. Further, computational model of EhODC dimer was generated. The homodimer contains two separate active sites at the dimer interface with Lys57 and Cys334 residues of opposite monomers contributing to each active site. Molecular dynamic simulations were performed and the dimeric structure was found to be very stable with RMSD value ∼0.327 nm. To gain insight into the functional role, the interface residues critical for dimerization and active site formation were identified and mutated. Mutation of Lys57Ala or Cys334Ala completely abolished enzyme activity. Interestingly, partial restoration of the enzyme activity was observed when inactive Lys57Ala and Cys334Ala mutants were mixed confirming that the dimer is the active form. Furthermore, Gly361Tyr and Lys157Ala mutations at the dimer interface were found to abolish the enzyme activity and destabilize the dimer.
Conclusion: To our knowledge, this is the first report which demonstrates that EhODC is functional in the dimeric form. These findings and availability of 3D structure model of EhODC dimer opens up possibilities for alternate enzyme inhibition strategies by targeting the dimer disruption.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




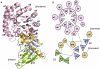

Similar articles
-
Structural insight into DFMO resistant ornithine decarboxylase from Entamoeba histolytica: an inkling to adaptive evolution.PLoS One. 2013;8(1):e53397. doi: 10.1371/journal.pone.0053397. Epub 2013 Jan 11. PLoS One. 2013. PMID: 23326423 Free PMC article.
-
Characterization of the Entamoeba histolytica ornithine decarboxylase-like enzyme.PLoS Negl Trop Dis. 2008 Jan 2;2(1):e115. doi: 10.1371/journal.pntd.0000115. PLoS Negl Trop Dis. 2008. PMID: 18235846 Free PMC article.
-
Entamoeba histolytica, heterologous expression and in situ immunolocalization of ornithine decarboxylase (EhODC).Exp Parasitol. 2009 Sep;123(1):99-104. doi: 10.1016/j.exppara.2009.06.005. Epub 2009 Jun 9. Exp Parasitol. 2009. PMID: 19520076
-
Entamoeba histolytica: purification and characterization of ornithine decarboxylase.Exp Parasitol. 2002 Aug;101(4):215-22. doi: 10.1016/s0014-4894(02)00137-6. Exp Parasitol. 2002. PMID: 12594962
-
Rational design of ornithine decarboxylase with high catalytic activity for the production of putrescine.Appl Microbiol Biotechnol. 2014 Sep;98(17):7483-90. doi: 10.1007/s00253-014-5669-8. Epub 2014 Apr 5. Appl Microbiol Biotechnol. 2014. PMID: 24706212
Cited by
-
Structural insight into DFMO resistant ornithine decarboxylase from Entamoeba histolytica: an inkling to adaptive evolution.PLoS One. 2013;8(1):e53397. doi: 10.1371/journal.pone.0053397. Epub 2013 Jan 11. PLoS One. 2013. PMID: 23326423 Free PMC article.
-
Proteomic Identification of Oxidized Proteins in Entamoeba histolytica by Resin-Assisted Capture: Insights into the Role of Arginase in Resistance to Oxidative Stress.PLoS Negl Trop Dis. 2016 Jan 6;10(1):e0004340. doi: 10.1371/journal.pntd.0004340. eCollection 2016 Jan. PLoS Negl Trop Dis. 2016. PMID: 26735309 Free PMC article.
-
N-acetyl ornithine deacetylase is a moonlighting protein and is involved in the adaptation of Entamoeba histolytica to nitrosative stress.Sci Rep. 2016 Nov 3;6:36323. doi: 10.1038/srep36323. Sci Rep. 2016. PMID: 27808157 Free PMC article.
References
-
- Rosas-Arreguín P, Arteaga-Nieto P, Reynoso-Orozco R, Villagómez-Castro JC, Sabanero-López M, et al. Bursera fagaroides, effect of an ethanolic extract on ornithine decarboxylase (ODC) activity in vitro and on the growth of Entamoeba histolytica. Exp Parasitol. 2008;119:398–402. - PubMed
-
- López-Vallejo F, Castillo R, Yépez-Mulia L, Medina-Franco JL. Benzotriazoles and indazoles are scaffolds with biological activity against Entamoeba histolytica. J Biomol Screen. 2011;16:862–868. - PubMed
-
- Petri WA., Jr Therapy of intestinal protozoa. Trends Parasitol. 2003;19:523–526. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

