ATRX-mediated chromatin association of histone variant macroH2A1 regulates α-globin expression
- PMID: 22391447
- PMCID: PMC3305981
- DOI: 10.1101/gad.179416.111
ATRX-mediated chromatin association of histone variant macroH2A1 regulates α-globin expression
Abstract
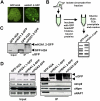
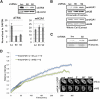
The histone variant macroH2A generally associates with transcriptionally inert chromatin; however, the factors that regulate its chromatin incorporation remain elusive. Here, we identify the SWI/SNF helicase ATRX (α-thalassemia/MR, X-linked) as a novel macroH2A-interacting protein. Unlike its role in assisting H3.3 chromatin deposition, ATRX acts as a negative regulator of macroH2A's chromatin association. In human erythroleukemic cells deficient for ATRX, macroH2A accumulates at the HBA gene cluster on the subtelomere of chromosome 16, coinciding with the loss of α-globin expression. Collectively, our results implicate deregulation of macroH2A's distribution as a contributing factor to the α-thalassemia phenotype of ATRX syndrome.
Figures
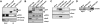
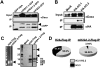
References
-
- Angelov D, Molla A, Perche PY, Hans F, Cote J, Khochbin S, Bouvet P, Dimitrov S 2003. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell 11: 1033–1041 - PubMed
-
- Berube NG 2011. ATRX in chromatin assembly and genome architecture during development and disease. Biochem Cell Biol 89: 435–444 - PubMed
-
- Berube NG, Smeenk CA, Picketts DJ 2000. Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum Mol Genet 9: 539–547 - PubMed
-
- Buschbeck M, Uribesalgo I, Wibowo I, Rue P, Martin D, Gutierrez A, Morey L, Guigo R, Lopez-Schier H, Di Croce L 2009. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat Struct Mol Biol 16: 1074–1079 - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
