Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography
- PMID: 22392984
- PMCID: PMC3311327
- DOI: 10.1073/pnas.1201333109
Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography
Abstract
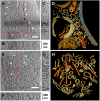
Cryoelectron tomography provides unprecedented insights into the macromolecular and supramolecular organization of cells in a close-to-living state. However because of the limited thickness range (< 0.5-1 μm) that is accessible with today's intermediate voltage electron microscopes only small prokaryotic cells or peripheral regions of eukaryotic cells can be examined directly. Key to overcoming this limitation is the ability to prepare sufficiently thin samples. Cryosectioning can be used to prepare thin enough sections but suffers from severe artefacts, such as substantial compression. Here we describe a procedure, based upon focused ion beam (FIB) milling for the preparation of thin (200-500 nm) lamellae from vitrified cells grown on electron microscopy (EM) grids. The self-supporting lamellae are apparently free of distortions or other artefacts and open up large windows into the cell's interior allowing tomographic studies to be performed on any chosen part of the cell. We illustrate the quality of sample preservation with a structure of the nuclear pore complex obtained from a single tomogram.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Comment in
-
Etch-a-cell.Nat Methods. 2012 May;9(5):434. doi: 10.1038/nmeth.2006. Nat Methods. 2012. PMID: 22803201 No abstract available.
References
-
- Koster AJ, et al. Perspectives of molecular and cellular electron tomography. J Struct Biol. 1997;120:276–308. - PubMed
-
- Lucic V, Forster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem. 2005;74:833–865. - PubMed
-
- Baumeister W. From proteomic inventory to architecture. FEBS Lett. 2005;579:933–937. - PubMed
-
- Grunewald K, et al. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. - PubMed
-
- Medalia O, et al. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298:1209–1213. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

