Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods
- PMID: 22393009
- PMCID: PMC3311382
- DOI: 10.1073/pnas.1200068109
Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods
Abstract
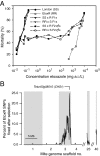
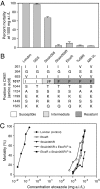
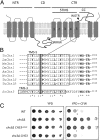
Because of its importance to the arthropod exoskeleton, chitin biogenesis is an attractive target for pest control. This point is demonstrated by the economically important benzoylurea compounds that are in wide use as highly specific agents to control insect populations. Nevertheless, the target sites of compounds that inhibit chitin biogenesis have remained elusive, likely preventing the full exploitation of the underlying mode of action in pest management. Here, we show that the acaricide etoxazole inhibits chitin biogenesis in Tetranychus urticae (the two-spotted spider mite), an economically important pest. We then developed a population-level bulk segregant mapping method, based on high-throughput genome sequencing, to identify a locus for monogenic, recessive resistance to etoxazole in a field-collected population. As supported by additional genetic studies, including sequencing across multiple resistant strains and genetic complementation tests, we associated a nonsynonymous mutation in the major T. urticae chitin synthase (CHS1) with resistance. The change is in a C-terminal transmembrane domain of CHS1 in a highly conserved region that may serve a noncatalytic but essential function. Our finding of a target-site resistance mutation in CHS1 shows that at least one highly specific chitin biosynthesis inhibitor acts directly to inhibit chitin synthase. Our work also raises the possibility that other chitin biogenesis inhibitors, such as the benzoylurea compounds, may also act by inhibition of chitin synthases. More generally, our genetic mapping approach should be powerful for high-resolution mapping of simple traits (resistance or otherwise) in arthropods.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Cohen E. Chitin biochemistry: Synthesis, hydrolysis and inhibition. In: Jérôme C, Stephen JS, editors. Adv Insect Physiol. Vol 38. London: Academic; 2010. pp. 5–74.
-
- Merzendorfer H. Insect chitin synthases: A review. J Comp Physiol B. 2006;176:1–15. - PubMed
-
- Ishaaya I, Casida JE. Dietary TH 6040 alters composition and enzyme-activity of housefly larval cuticle. Pestic Biochem Physiol. 1974;4:484–490.
-
- Palli SR, Retnakaran A. Molecular and biochemical aspects of chitin synthesis inhibition. EXS. 1999;87:85–98. - PubMed
-
- Verloop A, Ferrel CD. Benzoylphenyl ureas—A new group of larvicides interfering with chitin deposition. Pesticide Chemistry in the 20th Century. ACS Symposium Series. In: Plummer JR, editor; Plummer JR, editor. Vol 37. Washington, DC: Am Chem Soc; 1977. pp. 237–270.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

