A RUNX2-HDAC1 co-repressor complex regulates rRNA gene expression by modulating UBF acetylation
- PMID: 22393235
- PMCID: PMC3403236
- DOI: 10.1242/jcs.100909
A RUNX2-HDAC1 co-repressor complex regulates rRNA gene expression by modulating UBF acetylation
Abstract
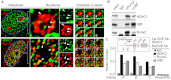
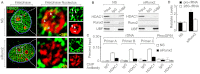
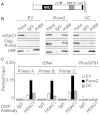
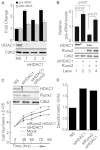
The osteogenic and oncogenic transcription factor RUNX2 downregulates the RNA polymerase I (RNA Pol I)-mediated transcription of rRNAs and changes histone modifications associated with the rDNA repeat. However, the mechanisms by which RUNX2 suppresses rRNA transcription are not well understood. RUNX2 cofactors such as histone deacetylases (HDACs) play a key role in chromatin remodeling and regulation of gene transcription. Here, we show that RUNX2 recruits HDAC1 to the rDNA repeats in osseous cells. This recruitment alters the histone modifications associated with active rRNA-encoding genes and causes deacetylation of the protein upstream binding factor (UBF, also known as UBTF). Downregulation of RUNX2 expression reduces the localization of HDAC1 to the nucleolar periphery and also decreases the association between HDAC1 and UBF. Functionally, depletion of HDAC1 relieves the RUNX2-mediated repression of rRNA-encoding genes and concomitantly increases cell proliferation and global protein synthesis in osseous cells. Our findings collectively identify a RUNX2-HDAC1-dependent mechanism for the regulation of rRNA-encoding genes and suggest that there is plasticity to RUNX2-mediated epigenetic control, which is mediated through selective mitotic exclusion of co-regulatory factors.
Figures
References
-
- Arabi A., Wu S., Ridderstråle K., Bierhoff H., Shiue C., Fatyol K., Fahlén S., Hydbring P., Söderberg O., Grummt I., et al. (2005). c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 7, 303-310 - PubMed
-
- Bell S. P., Learned R. M., Jantzen H. M., Tjian R. (1988). Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 241, 1192-1197 - PubMed
-
- Budde A., Grummt I. (1999). p53 represses ribosomal gene transcription. Oncogene 18, 1119-1124 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

