Single-molecule tools elucidate H2A.Z nucleosome composition
- PMID: 22393239
- PMCID: PMC3434807
- DOI: 10.1242/jcs.101592
Single-molecule tools elucidate H2A.Z nucleosome composition
Abstract
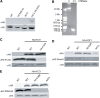
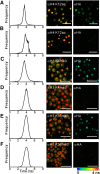
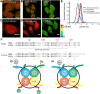
Although distinct epigenetic marks correlate with different chromatin states, how they are integrated within single nucleosomes to generate combinatorial signals remains largely unknown. We report the successful implementation of single molecule tools constituting fluorescence correlation spectroscopy (FCS), pulse interleave excitation-based Förster resonance energy transfer (PIE-FRET) and fluorescence lifetime imaging-based FRET (FLIM-FRET) to elucidate the composition of single nucleosomes containing histone variant H2A.Z (Htz1p in yeast) in vitro and in vivo. We demonstrate that yeast nucleosomes containing Htz1p are primarily composed of H4 K12ac and H3 K4me3 but not H3 K36me3 and that these patterns are conserved in mammalian cells. Quantification of epigenetic modifications in nucleosomes will provide a new dimension to epigenetics research and lead to a better understanding of how these patterns contribute to the targeting of chromatin-binding proteins and chromatin structure during gene regulation.
Figures

Similar articles
-
Global dynamics of newly constructed oligonucleosomes of conventional and variant H2A.Z histone.BMC Struct Biol. 2007 Nov 8;7:76. doi: 10.1186/1472-6807-7-76. BMC Struct Biol. 2007. PMID: 17996059 Free PMC article.
-
Splitting of H3-H4 tetramers at transcriptionally active genes undergoing dynamic histone exchange.Proc Natl Acad Sci U S A. 2011 Jan 25;108(4):1296-301. doi: 10.1073/pnas.1018308108. Epub 2011 Jan 10. Proc Natl Acad Sci U S A. 2011. PMID: 21220302 Free PMC article.
-
Cryo-EM structures reveal the acetylation process of piccolo NuA4.Proc Natl Acad Sci U S A. 2025 Mar 25;122(12):e2414490122. doi: 10.1073/pnas.2414490122. Epub 2025 Mar 18. Proc Natl Acad Sci U S A. 2025. PMID: 40100634
-
The specificity of H2A.Z occupancy in the yeast genome and its relationship to transcription.Curr Genet. 2020 Oct;66(5):939-944. doi: 10.1007/s00294-020-01087-7. Epub 2020 Jun 14. Curr Genet. 2020. PMID: 32537667 Review.
-
Contributions of Histone Variants in Nucleosome Structure and Function.J Mol Biol. 2021 Mar 19;433(6):166678. doi: 10.1016/j.jmb.2020.10.012. Epub 2020 Oct 14. J Mol Biol. 2021. PMID: 33065110 Review.
Cited by
-
Optogenetic regulation of site-specific subtelomeric DNA methylation.Oncotarget. 2016 Aug 2;7(31):50380-50391. doi: 10.18632/oncotarget.10394. Oncotarget. 2016. PMID: 27391261 Free PMC article.
-
Embedding a membrane protein into an enveloped artificial viral replica.RSC Chem Biol. 2021 Dec 21;3(2):231-241. doi: 10.1039/d1cb00166c. eCollection 2022 Feb 9. RSC Chem Biol. 2021. PMID: 35360888 Free PMC article.
-
Molecular probes for fluorescence lifetime imaging.Bioconjug Chem. 2015 Jun 17;26(6):963-74. doi: 10.1021/acs.bioconjchem.5b00167. Epub 2015 May 22. Bioconjug Chem. 2015. PMID: 25961514 Free PMC article. Review.
-
Global nutrition research: nutrition and breast cancer prevention as a model.Nutr Rev. 2013 Nov;71(11):742-52. doi: 10.1111/nure.12075. Epub 2013 Oct 22. Nutr Rev. 2013. PMID: 24447199 Free PMC article. Review.
-
Monitoring focal adhesion kinase phosphorylation dynamics in live cells.Analyst. 2017 Jul 24;142(15):2713-2716. doi: 10.1039/c7an00471k. Analyst. 2017. PMID: 28589989 Free PMC article.
References
-
- Adams A., Gottschling D. E., Kaiser C. A., Stearns T. (1997). Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press
-
- Allard S., Utley R. T., Savard J., Clarke A., Grant P., Brandl C. J., Pillus L., Workman J. L., Côté J. (1999). NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18, 5108–5119 10.1093/emboj/18.18.5108 - DOI - PMC - PubMed
-
- Altaf M., Auger A., Monnet–Saksouk J., Brodeur J., Piquet S., Cramet M., Bouchard N., Lacoste N., Utley R. T., Gaudreau L., et al. (2010). NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J. Biol. Chem. 285, 15966–15977 10.1074/jbc.M110.117069 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

