Epigenetic silencing of HNF1A associates with changes in the composition of the human plasma N-glycome
- PMID: 22395466
- PMCID: PMC3335910
- DOI: 10.4161/epi.7.2.18918
Epigenetic silencing of HNF1A associates with changes in the composition of the human plasma N-glycome
Abstract
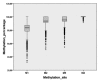
Protein glycosylation is a ubiquitous modification that affects the structure and function of proteins. Our recent genome wide association study identified transcription factor HNF1A as an important regulator of plasma protein glycosylation. To evaluate the potential impact of epigenetic regulation of HNF1A on protein glycosylation we analyzed CpG methylation in 810 individuals. The association between methylation of four CpG sites and the composition of plasma and IgG glycomes was analyzed. Several statistically significant associations were observed between HNF1A methylation and plasma glycans, while there were no significant associations with IgG glycans. The most consistent association with HNF1A methylation was observed with the increase in the proportion of highly branched glycans in the plasma N-glycome. The hypothesis that inactivation of HNF1A promotes branching of glycans was supported by the analysis of plasma N-glycomes in 61 patients with inactivating mutations in HNF1A, where the increase in plasma glycan branching was also observed. This study represents the first demonstration of epigenetic regulation of plasma glycome composition, suggesting a potential mechanism by which epigenetic deregulation of the glycome may contribute to disease development.
Figures
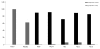


References
-
- Vaissière T, Hung RJ, Zaridze D, Moukeria A, Cuenin C, Fasolo V, et al. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res. 2009;69:243–252. doi: 10.1158/0008-5472.CAN-08-2489. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
