The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops
- PMID: 22395476
- PMCID: PMC3321525
- DOI: 10.1038/msb.2012.6
The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops
Abstract
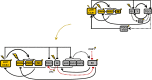
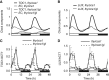
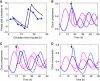
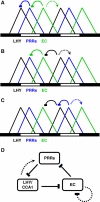
Circadian clocks synchronise biological processes with the day/night cycle, using molecular mechanisms that include interlocked, transcriptional feedback loops. Recent experiments identified the evening complex (EC) as a repressor that can be essential for gene expression rhythms in plants. Integrating the EC components in this role significantly alters our mechanistic, mathematical model of the clock gene circuit. Negative autoregulation of the EC genes constitutes the clock's evening loop, replacing the hypothetical component Y. The EC explains our earlier conjecture that the morning gene Pseudo-Response Regulator 9 was repressed by an evening gene, previously identified with Timing Of CAB Expression1 (TOC1). Our computational analysis suggests that TOC1 is a repressor of the morning genes Late Elongated Hypocotyl and Circadian Clock Associated1 rather than an activator as first conceived. This removes the necessity for the unknown component X (or TOC1mod) from previous clock models. As well as matching timeseries and phase-response data, the model provides a new conceptual framework for the plant clock that includes a three-component repressilator circuit in its complex structure.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 - PubMed
-
- Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D, Deng XW (2010) Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/D019621/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F005237/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G19886/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F005296/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E015263/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

