Genetic dating indicates that the Asian-Papuan admixture through Eastern Indonesia corresponds to the Austronesian expansion
- PMID: 22396590
- PMCID: PMC3311340
- DOI: 10.1073/pnas.1118892109
Genetic dating indicates that the Asian-Papuan admixture through Eastern Indonesia corresponds to the Austronesian expansion
Abstract
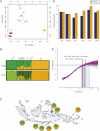
Although the Austronesian expansion had a major impact on the languages of Island Southeast Asia, controversy still exists over the genetic impact of this expansion. The coexistence of both Asian and Papuan genetic ancestry in Eastern Indonesia provides a unique opportunity to address this issue. Here, we estimate recombination breakpoints in admixed genomes based on genome-wide SNP data and date the genetic admixture between populations of Asian vs. Papuan ancestry in Eastern Indonesia. Analyses of two genome-wide datasets indicate an eastward progression of the Asian admixture signal in Eastern Indonesia beginning about 4,000-3,000 y ago, which is in excellent agreement with inferences based on Austronesian languages. The average rate of spread of Asian genes in Eastern Indonesia was about 0.9 km/y. Our results indicate that the Austronesian expansion had a strong genetic as well as linguistic impact on Island Southeast Asia, and they significantly advance our understanding of the biological origins of human populations in the Asia-Pacific region.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Lack of correspondence between Asian-Papuan genetic admixture and Austronesian language dispersal in eastern Indonesia.Proc Natl Acad Sci U S A. 2012 Sep 25;109(39):E2577; author reply E2578. doi: 10.1073/pnas.1209616109. Epub 2012 Aug 6. Proc Natl Acad Sci U S A. 2012. PMID: 22869694 Free PMC article. No abstract available.
References
-
- Bellwood PS, Fox JJ, Tryon DT. The Austronesians: Historical and Comparative Perspectives. Canberra, Australia: ANU E Press; 1995.
-
- Soares P, et al. Climate change and postglacial human dispersals in southeast Asia. Mol Biol Evol. 2008;25:1209–1218. - PubMed
-
- Donohue M, Denham T. Farming and language in Island Southeast Asia: Reframing Austronesian history. Curr Anthropol. 2010;51:223–256.
-
- Karafet TM, et al. Major east-west division underlies Y chromosome stratification across Indonesia. Mol Biol Evol. 2010;27:1833–1844. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

