Visualization and measurement of ATP levels in living cells replicating hepatitis C virus genome RNA
- PMID: 22396648
- PMCID: PMC3291659
- DOI: 10.1371/journal.ppat.1002561
Visualization and measurement of ATP levels in living cells replicating hepatitis C virus genome RNA
Abstract
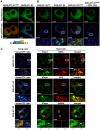
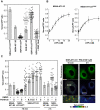
Adenosine 5'-triphosphate (ATP) is the primary energy currency of all living organisms and participates in a variety of cellular processes. Although ATP requirements during viral lifecycles have been examined in a number of studies, a method by which ATP production can be monitored in real-time, and by which ATP can be quantified in individual cells and subcellular compartments, is lacking, thereby hindering studies aimed at elucidating the precise mechanisms by which viral replication energized by ATP is controlled. In this study, we investigated the fluctuation and distribution of ATP in cells during RNA replication of the hepatitis C virus (HCV), a member of the Flaviviridae family. We demonstrated that cells involved in viral RNA replication actively consumed ATP, thereby reducing cytoplasmic ATP levels. Subsequently, a method to measure ATP levels at putative subcellular sites of HCV RNA replication in living cells was developed by introducing a recently-established Förster resonance energy transfer (FRET)-based ATP indicator, called ATeam, into the NS5A coding region of the HCV replicon. Using this method, we were able to observe the formation of ATP-enriched dot-like structures, which co-localize with non-structural viral proteins, within the cytoplasm of HCV-replicating cells but not in non-replicating cells. The obtained FRET signals allowed us to estimate ATP concentrations within HCV replicating cells as ∼5 mM at possible replicating sites and ∼1 mM at peripheral sites that did not appear to be involved in HCV replication. In contrast, cytoplasmic ATP levels in non-replicating Huh-7 cells were estimated as ∼2 mM. To our knowledge, this is the first study to demonstrate changes in ATP concentration within cells during replication of the HCV genome and increased ATP levels at distinct sites within replicating cells. ATeam may be a powerful tool for the study of energy metabolism during replication of the viral genome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
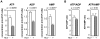



References
-
- Kato-Yamada Y, Yoshida M. Isolated epsilon subunit of thermophilic F1-ATPase binds ATP. J Biol Chem. 2003;278:36013–36016. - PubMed
-
- Iino R, Murakami T, Iizuka S, Kato-Yamada Y, Suzuki T, et al. Real-time monitoring of conformational dynamics of the epsilon subunit in F1-ATPase. J Biol Chem. 2005;280:40130–40134. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

