New connections between splicing and human disease
- PMID: 22397991
- PMCID: PMC3319163
- DOI: 10.1016/j.tig.2012.01.001
New connections between splicing and human disease
Abstract
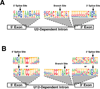
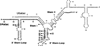
The removal by splicing of introns from the primary transcripts of most mammalian genes is an essential step in gene expression. Splicing is performed by large, complex ribonucleoprotein particles termed spliceosomes. Mammals contain two types that splice out mutually exclusive types of introns. However, the role of the minor spliceosome has been poorly studied. Recent reports have now shown that mutations in one minor spliceosomal snRNA, U4atac, are linked to a rare autosomal recessive developmental defect. In addition, very exciting recent results of exome deep-sequencing have found that recurrent, somatic, heterozygous mutations of other splicing factors occur at high frequencies in particular cancers and pre-cancerous conditions, suggesting that alterations in the core splicing machinery can contribute to tumorigenesis. Mis-splicing of crucial genes may underlie the pathologies of all of these diseases. Identifying these genes and understanding the mechanisms involved in their mis-splicing may lead to advancements in diagnosis and treatment.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
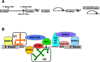

References
-
- Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. - PubMed
-
- Hall SL, Padgett RA. Conserved sequences in a class of rare eukaryotic introns with non-consensus splice sites. J. Mol. Biol. 1994;239:357–365. - PubMed
-
- Hall SL, Padgett RA. Requirement of U12 snRNA for the in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science. 1996;271:1716–1718. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

