The effect of geographical scale of sampling on DNA barcoding
- PMID: 22398121
- PMCID: PMC3417044
- DOI: 10.1093/sysbio/sys037
The effect of geographical scale of sampling on DNA barcoding
Abstract
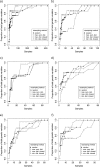
Eight years after DNA barcoding was formally proposed on a large scale, CO1 sequences are rapidly accumulating from around the world. While studies to date have mostly targeted local or regional species assemblages, the recent launch of the global iBOL project (International Barcode of Life), highlights the need to understand the effects of geographical scale on Barcoding's goals. Sampling has been central in the debate on DNA Barcoding, but the effect of the geographical scale of sampling has not yet been thoroughly and explicitly tested with empirical data. Here, we present a CO1 data set of aquatic predaceous diving beetles of the tribe Agabini, sampled throughout Europe, and use it to investigate how the geographic scale of sampling affects 1) the estimated intraspecific variation of species, 2) the genetic distance to the most closely related heterospecific, 3) the ratio of intraspecific and interspecific variation, 4) the frequency of taxonomically recognized species found to be monophyletic, and 5) query identification performance based on 6 different species assignment methods. Intraspecific variation was significantly correlated with the geographical scale of sampling (R-square = 0.7), and more than half of the species with 10 or more sampled individuals (N = 29) showed higher intraspecific variation than 1% sequence divergence. In contrast, the distance to the closest heterospecific showed a significant decrease with increasing geographical scale of sampling. The average genetic distance dropped from > 7% for samples within 1 km, to < 3.5% for samples up to > 6000 km apart. Over a third of the species were not monophyletic, and the proportion increased through locally, nationally, regionally, and continentally restricted subsets of the data. The success of identifying queries decreased with increasing spatial scale of sampling; liberal methods declined from 100% to around 90%, whereas strict methods dropped to below 50% at continental scales. The proportion of query identifications considered uncertain (more than one species < 1% distance from query) escalated from zero at local, to 50% at continental scale. Finally, by resampling the most widely sampled species we show that even if samples are collected to maximize the geographical coverage, up to 70 individuals are required to sample 95% of intraspecific variation. The results show that the geographical scale of sampling has a critical impact on the global application of DNA barcoding. Scale-effects result from the relative importance of different processes determining the composition of regional species assemblages (dispersal and ecological assembly) and global clades (demography, speciation, and extinction). The incorporation of geographical information, where available, will be required to obtain identification rates at global scales equivalent to those in regional barcoding studies. Our result hence provides an impetus for both smarter barcoding tools and sprouting national barcoding initiatives-smaller geographical scales deliver higher accuracy.
Figures









References
-
- Abdo Z, Golding B. A step toward barcoding life: a model-based, decision-theoretic method to assign genes to preexisting species groups. Syst. Biol. 2007;56:44–56. - PubMed
-
- Avise JC. Cambridge (MA): Harvard University Press; 2000. Phylogeography: the history and formation of species; p. 447.
-
- Ayoub NA, Riechert SE, Small RL. Speciation history of the North American funnel web spiders, Agelenopsis (Araneae: Agelenidae): phylogenetic inferences at the population–species interface. Mol. Phylogenet. Evol. 2005;36:42–57. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

