Plasticity and virus specificity of the airway epithelial cell immune response during respiratory virus infection
- PMID: 22398282
- PMCID: PMC3347264
- DOI: 10.1128/JVI.06757-11
Plasticity and virus specificity of the airway epithelial cell immune response during respiratory virus infection
Abstract
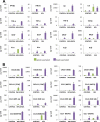
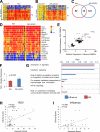
Airway epithelial cells (AECs) provide the first line of defense in the respiratory tract and are the main target of respiratory viruses. Here, using oligonucleotide and protein arrays, we analyze the infection of primary polarized human AEC cultures with influenza virus and respiratory syncytial virus (RSV), and we show that the immune response of AECs is quantitatively and qualitatively virus specific. Differentially expressed genes (DEGs) specifically induced by influenza virus and not by RSV included those encoding interferon B1 (IFN-B1), type III interferons (interleukin 28A [IL-28A], IL-28B, and IL-29), interleukins (IL-6, IL-1A, IL-1B, IL-23A, IL-17C, and IL-32), and chemokines (CCL2, CCL8, and CXCL5). Lack of type I interferon or STAT1 signaling decreased the expression and secretion of cytokines and chemokines by the airway epithelium. We also observed strong basolateral polarization of the secretion of cytokines and chemokines by human and murine AECs during infection. Importantly, the antiviral response of human AECs to influenza virus or to RSV correlated with the infection signature obtained from peripheral blood mononuclear cells (PBMCs) isolated from patients with acute influenza or RSV bronchiolitis, respectively. IFI27 (also known as ISG12) was identified as a biomarker of respiratory virus infection in both AECs and PBMCs. In addition, the extent of the transcriptional perturbation in PBMCs correlated with the clinical disease severity. Our results demonstrate that the human airway epithelium mounts virus-specific immune responses that are likely to determine the subsequent systemic immune responses and suggest that the absence of epithelial immune mediators after RSV infection may contribute to explaining the inadequacy of systemic immunity to the virus.
Figures





References
-
- Bals R, Hiemstra PS. 2004. Innate immunity in the lung: how epithelial cells fight against respiratory pathogens. Eur. Respir. J. 23:327–333 - PubMed
-
- Bernasconi D, Amici C, La Frazia S, Ianaro A, Santoro MG. 2005. The IkappaB kinase is a key factor in triggering influenza A virus-induced inflammatory cytokine production in airway epithelial cells. J. Biol. Chem. 280:24127–24134 - PubMed
-
- Bertrand P, Aranibar H, Castro E, Sanchez I. 2001. Efficacy of nebulized epinephrine versus salbutamol in hospitalized infants with bronchiolitis. Pediatr. Pulmonol. 31:284–288 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

