Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination
- PMID: 22398290
- PMCID: PMC3347312
- DOI: 10.1128/JVI.00146-12
Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination
Abstract
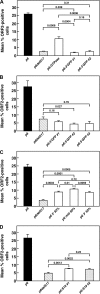
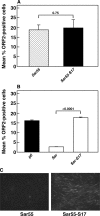
An infectious cDNA clone of a genotype 3 strain of hepatitis E virus adapted to growth in HepG2/C3A human hepatoma cells was constructed. This virus was unusual in that the hypervariable region of the adapted virus contained a 171-nucleotide insertion that encoded 58 amino acids of human S17 ribosomal protein. Analyses of virus from six serial passages indicated that genomes with this insert, although initially rare, were selected during the first passage, suggesting it conferred a significant growth advantage. RNA transcripts from this cDNA and the viruses encoded by them were infectious for cells of both human and swine origin, the major host species for this zoonotic virus. Mutagenesis studies demonstrated that the S17 insert was a major factor in cell culture adaptation. Introduction of 54 synonymous mutations into the insert had no detectable effect, thus implicating protein, rather than RNA, as the important component. Truncation of the insert by 50% decreased the levels of successful transfection by ~3-fold. Substitution of the S17 sequence by a different ribosomal protein sequence or by GTPase-activating protein sequence resulted in a partial enhancement of transfection levels, whereas substitution with 58 amino acids of green fluorescent protein had no effect. Therefore, both the sequence length and the amino acid composition of the insert were important. The S17 sequence did not affect transfection of human hepatoma cells when inserted into the hypervariable region of a genotype 1 strain, but this chimeric genome acquired a dramatic ability to replicate in hamster cells.
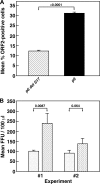
Figures




References
-
- Ahmad I, Holla RP, Jameel S. 2011. Molecular virology of hepatitis E virus. Virus Res. doi:10.1016/j.virusres.2011.02.11 - DOI - PMC - PubMed
-
- Dalton HR, Bendall RP, Keane FE, Tedder RS, Ijaz S. 2009. Persistent carriage of hepatitis E virus in patients with HIV infection. N. Engl. J. Med. 361:1025–1027 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

