Detecting and annotating genetic variations using the HugeSeq pipeline
- PMID: 22398614
- PMCID: PMC4720384
- DOI: 10.1038/nbt.2134
Detecting and annotating genetic variations using the HugeSeq pipeline
Conflict of interest statement
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at
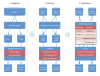
Figures

References
-
- Dean J, Ghemawat S. MapReduce: simplified data processing on large clusters. OSDI’04 Proceedings of the 6th Symposium on Operating Systems Design and Implementation; San Francisco. 2004.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

