Rab11-FIP3 is a cell cycle-regulated phosphoprotein
- PMID: 22401586
- PMCID: PMC3310825
- DOI: 10.1186/1471-2121-13-4
Rab11-FIP3 is a cell cycle-regulated phosphoprotein
Abstract
Background: Rab11 and its effector molecule, Rab11-FIP3 (FIP3), associate with recycling endosomes and traffic into the furrow and midbody of cells during cytokinesis. FIP3 also controls recycling endosome distribution during interphase. Here, we examine whether phosphorylation of FIP3 is involved in these activities.
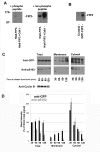
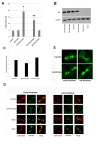
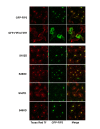
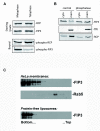
Results: We identify four sites of phosphorylation of FIP3 in vivo, S-102, S-280, S-347 and S-450 and identify S-102 as a target for Cdk1-cyclin B in vitro. Of these, we show that S-102 is phosphorylated in metaphase and is dephosphorylated as cells enter telophase. Over-expression of FIP3-S102D increased the frequency of binucleate cells consistent with a role for this phospho-acceptor site in cytokinesis. Mutation of S-280, S-347 or S-450 or other previously identified phospho-acceptor sites (S-488, S-538, S-647 and S-648) was without effect on binucleate cell formation and did not modulate the distribution of FIP3 during the cell cycle. In an attempt to identify a functional role for FIP3 phosphorylation, we report that the change in FIP3 distribution from cytosolic to membrane-associated observed during progression from anaphase to telophase is accompanied by a concomitant dephosphorylation of FIP3. However, the phospho-acceptor sites identified here did not control this change in distribution.
Conclusions: Our data thus identify FIP3 as a cell cycle regulated phosphoprotein and suggest dephosphorylation of FIP3 accompanies its translocation from the cytosol to membranes during telophase. S102 is dephosphorylated during telophase; mutation of S102 exerts a modest effect on cytokinesis. Finally, we show that de/phosphorylation of the phospho-acceptor sites identified here (S-102, S-280, S-347 and S-450) is not required for the spatial control of recycling endosome distribution or function.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

