Atomic view of a toxic amyloid small oligomer
- PMID: 22403391
- PMCID: PMC3959867
- DOI: 10.1126/science.1213151
Atomic view of a toxic amyloid small oligomer
Abstract
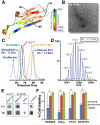
Amyloid diseases, including Alzheimer's, Parkinson's, and the prion conditions, are each associated with a particular protein in fibrillar form. These amyloid fibrils were long suspected to be the disease agents, but evidence suggests that smaller, often transient and polymorphic oligomers are the toxic entities. Here, we identify a segment of the amyloid-forming protein αB crystallin, which forms an oligomeric complex exhibiting properties of other amyloid oligomers: β-sheet-rich structure, cytotoxicity, and recognition by an oligomer-specific antibody. The x-ray-derived atomic structure of the oligomer reveals a cylindrical barrel, formed from six antiparallel protein strands, that we term a cylindrin. The cylindrin structure is compatible with a sequence segment from the β-amyloid protein of Alzheimer's disease. Cylindrins offer models for the hitherto elusive structures of amyloid oligomers.
Figures

Comment in
-
Biochemistry. Visualizing amyloid assembly.Science. 2012 Apr 20;336(6079):308-9. doi: 10.1126/science.1220356. Science. 2012. PMID: 22517851 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

