Sulfate-reducing microorganisms in wetlands - fameless actors in carbon cycling and climate change
- PMID: 22403575
- PMCID: PMC3289269
- DOI: 10.3389/fmicb.2012.00072
Sulfate-reducing microorganisms in wetlands - fameless actors in carbon cycling and climate change
Abstract
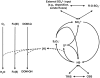
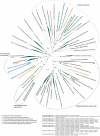
Freshwater wetlands are a major source of the greenhouse gas methane but at the same time can function as carbon sink. Their response to global warming and environmental pollution is one of the largest unknowns in the upcoming decades to centuries. In this review, we highlight the role of sulfate-reducing microorganisms (SRM) in the intertwined element cycles of wetlands. Although regarded primarily as methanogenic environments, biogeochemical studies have revealed a previously hidden sulfur cycle in wetlands that can sustain rapid renewal of the small standing pools of sulfate. Thus, dissimilatory sulfate reduction, which frequently occurs at rates comparable to marine surface sediments, can contribute up to 36-50% to anaerobic carbon mineralization in these ecosystems. Since sulfate reduction is thermodynamically favored relative to fermentative processes and methanogenesis, it effectively decreases gross methane production thereby mitigating the flux of methane to the atmosphere. However, very little is known about wetland SRM. Molecular analyses using dsrAB [encoding subunit A and B of the dissimilatory (bi)sulfite reductase] as marker genes demonstrated that members of novel phylogenetic lineages, which are unrelated to recognized SRM, dominate dsrAB richness and, if tested, are also abundant among the dsrAB-containing wetland microbiota. These discoveries point toward the existence of so far unknown SRM that are an important part of the autochthonous wetland microbiota. In addition to these numerically dominant microorganisms, a recent stable isotope probing study of SRM in a German peatland indicated that rare biosphere members might be highly active in situ and have a considerable stake in wetland sulfate reduction. The hidden sulfur cycle in wetlands and the fact that wetland SRM are not well represented by described SRM species explains their so far neglected role as important actors in carbon cycling and climate change.
Keywords: dissimilatory (bi)sulfite reductase; dsrAB; peatland; rare biosphere; rice paddy; sulfate-reducing microorganisms; sulfur cycle; sulfur pollution.
Figures


References
-
- Achtnich C., Bak F., Conrad R. (1995). Competition for electron donors among nitrate reducers, ferric iron reducers, sulfate reducers, and methanogens in anoxic paddy soil. Biol. Fertil. Soils 19, 65–72 10.1007/BF00336349 - DOI
-
- Alewell C., Gehre M. (1999). Patterns of stable S isotopes in a forested catchment as indicators for biological S turnover. Biogeochemistry 47, 319–333 10.1023/A:1006149804696 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

