Detecting remote sequence homology in disordered proteins: discovery of conserved motifs in the N-termini of Mononegavirales phosphoproteins
- PMID: 22403617
- PMCID: PMC3293882
- DOI: 10.1371/journal.pone.0031719
Detecting remote sequence homology in disordered proteins: discovery of conserved motifs in the N-termini of Mononegavirales phosphoproteins
Abstract
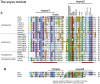
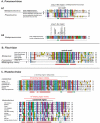
Paramyxovirinae are a large group of viruses that includes measles virus and parainfluenza viruses. The viral Phosphoprotein (P) plays a central role in viral replication. It is composed of a highly variable, disordered N-terminus and a conserved C-terminus. A second viral protein alternatively expressed, the V protein, also contains the N-terminus of P, fused to a zinc finger. We suspected that, despite their high variability, the N-termini of P/V might all be homologous; however, using standard approaches, we could previously identify sequence conservation only in some Paramyxovirinae. We now compared the N-termini using sensitive sequence similarity search programs, able to detect residual similarities unnoticeable by conventional approaches. We discovered that all Paramyxovirinae share a short sequence motif in their first 40 amino acids, which we called soyuz1. Despite its short length (11-16aa), several arguments allow us to conclude that soyuz1 probably evolved by homologous descent, unlike linear motifs. Conservation across such evolutionary distances suggests that soyuz1 plays a crucial role and experimental data suggest that it binds the viral nucleoprotein to prevent its illegitimate self-assembly. In some Paramyxovirinae, the N-terminus of P/V contains a second motif, soyuz2, which might play a role in blocking interferon signaling. Finally, we discovered that the P of related Mononegavirales contain similarly overlooked motifs in their N-termini, and that their C-termini share a previously unnoticed structural similarity suggesting a common origin. Our results suggest several testable hypotheses regarding the replication of Mononegavirales and suggest that disordered regions with little overall sequence similarity, common in viral and eukaryotic proteins, might contain currently overlooked motifs (intermediate in length between linear motifs and disordered domains) that could be detected simply by comparing orthologous proteins.
Conflict of interest statement
Figures


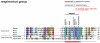





References
-
- Whelan SP, Barr JN, Wertz GW. Transcription and replication of nonsegmented negative-strand RNA viruses. Current Topics in Microbiology and Immunology. 2004;283:61–119. - PubMed
-
- Fontana JM, Bankamp B, Rota PA. Inhibition of interferon induction and signaling by paramyxoviruses. Immunological Reviews. 2008;225:46–67. - PubMed
-
- Habchi J, Longhi S. Structural disorder within paramyxovirus nucleoproteins and phosphoproteins. Molecular Biosystems. 2011;8:69–81. - PubMed
-
- Leyrat C, Gerard FCA, Ribeiro ED, Ivanov I, Ruigrok RWH, et al. Structural disorder in proteins of the rhabdoviridae replication complex. Protein and Peptide Letters. 2010;17:979–987. - PubMed
-
- Karlin D, Ferron F, Canard B, Longhi S. Structural disorder and modular organization in paramyxovirinae N and P. Journal of General Virology. 2003;84:3239–3252. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

