Isolation of a 97-kb minimal essential MHC B locus from a new reverse-4D BAC library of the golden pheasant
- PMID: 22403630
- PMCID: PMC3293878
- DOI: 10.1371/journal.pone.0032154
Isolation of a 97-kb minimal essential MHC B locus from a new reverse-4D BAC library of the golden pheasant
Abstract
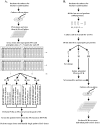
The bacterial artificial chromosome (BAC) system is widely used in isolation of large genomic fragments of interest. Construction of a routine BAC library requires several months for picking clones and arraying BACs into superpools in order to employ 4D-PCR to screen positive BACs, which might be time-consuming and laborious. The major histocompatibility complex (MHC) is a cluster of genes involved in the vertebrate immune system, and the classical avian MHC-B locus is a minimal essential one, occupying a 100-kb genomic region. In this study, we constructed a more effective reverse-4D BAC library for the golden pheasant, which first creates sub-libraries and then only picks clones of positive sub-libraries, and identified several MHC clones within thirty days. The full sequencing of a 97-kb reverse-4D BAC demonstrated that the golden pheasant MHC-B locus contained 20 genes and showed good synteny with that of the chicken. The notable differences between these two species were the numbers of class II B loci and NK genes and the inversions of the TAPBP gene and the TAP1-TAP2 region. Furthermore, the inverse TAP2-TAP1 was unique in the golden pheasant in comparison with that of chicken, turkey, and quail. The newly defined genomic structure of the golden pheasant MHC will give an insight into the evolutionary history of the avian MHC.
Conflict of interest statement
Figures



Similar articles
-
Sequencing of the core MHC region of black grouse (Tetrao tetrix) and comparative genomics of the galliform MHC.BMC Genomics. 2012 Oct 15;13:553. doi: 10.1186/1471-2164-13-553. BMC Genomics. 2012. PMID: 23066932 Free PMC article.
-
A high-density BAC physical map covering the entire MHC region of addax antelope genome.BMC Genomics. 2019 Jun 11;20(1):479. doi: 10.1186/s12864-019-5790-2. BMC Genomics. 2019. PMID: 31185912 Free PMC article.
-
Historical gene flow and profound spatial genetic structure among golden pheasant populations suggested by multi-locus analysis.Mol Phylogenet Evol. 2017 May;110:93-103. doi: 10.1016/j.ympev.2017.03.009. Epub 2017 Mar 7. Mol Phylogenet Evol. 2017. PMID: 28286102
-
Molecular characterization of classical and nonclassical MHC class I genes from the golden pheasant (Chrysolophus pictus).Int J Immunogenet. 2016 Feb;43(1):8-17. doi: 10.1111/iji.12245. Epub 2015 Dec 24. Int J Immunogenet. 2016. PMID: 26700854
-
Giant panda BAC library construction and assembly of a 650-kb contig spanning major histocompatibility complex class II region.BMC Genomics. 2007 Sep 8;8:315. doi: 10.1186/1471-2164-8-315. BMC Genomics. 2007. PMID: 17825108 Free PMC article.
Cited by
-
Molecular Characterization of MHC Class I Genes in Four Species of the Turdidae Family to Assess Genetic Diversity and Selection.Biomed Res Int. 2021 Apr 10;2021:5585687. doi: 10.1155/2021/5585687. eCollection 2021. Biomed Res Int. 2021. PMID: 33937397 Free PMC article.
-
Characterization of major histocompatibility complex class I loci of the lark sparrow (Chondestes grammacus) and insights into avian MHC evolution.Genetica. 2015 Aug;143(4):521-34. doi: 10.1007/s10709-015-9850-5. Epub 2015 Jun 13. Genetica. 2015. PMID: 26071093
-
Sequencing of the core MHC region of black grouse (Tetrao tetrix) and comparative genomics of the galliform MHC.BMC Genomics. 2012 Oct 15;13:553. doi: 10.1186/1471-2164-13-553. BMC Genomics. 2012. PMID: 23066932 Free PMC article.
-
Reconstructing Macroevolutionary Patterns in Avian MHC Architecture With Genomic Data.Front Genet. 2022 Feb 17;13:823686. doi: 10.3389/fgene.2022.823686. eCollection 2022. Front Genet. 2022. PMID: 35251132 Free PMC article.
-
Comparative genome analyses reveal distinct structure in the saltwater crocodile MHC.PLoS One. 2014 Dec 11;9(12):e114631. doi: 10.1371/journal.pone.0114631. eCollection 2014. PLoS One. 2014. PMID: 25503521 Free PMC article.
References
-
- Miyake T, Amemiya CT. BAC libraries and comparative genomics of aquatic chordate species. Comparative Biochemistry and Physiology Part C: Toxicology & Pharmacology. 2004;138:233–244. - PubMed
-
- Kim UJ, Birren BW, Slepak T, Mancino V, Boysen C, et al. Construction and characterization of a human bacterial artificial chromosome library. Genomics. 1996;34:213–218. - PubMed
-
- Chaves LD, Krueth SB, Reed KM. Defining the turkey MHC: sequence and genes of the B locus. The Journal of Immunology. 2009;183:6530. - PubMed
-
- Klein J. Natural history of the major histocompatibility complex. New York: Wiley; 1986.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

