RNA polymerase II elongation control
- PMID: 22404626
- PMCID: PMC4273853
- DOI: 10.1146/annurev-biochem-052610-095910
RNA polymerase II elongation control
Abstract
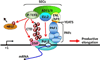
Regulation of the elongation phase of transcription by RNA polymerase II (Pol II) is utilized extensively to generate the pattern of mRNAs needed to specify cell types and to respond to environmental changes. After Pol II initiates, negative elongation factors cause it to pause in a promoter proximal position. These polymerases are poised to respond to the positive transcription elongation factor P-TEFb, and then enter productive elongation only under the appropriate set of signals to generate full-length properly processed mRNAs. Recent global analyses of Pol II and elongation factors, mechanisms that regulate P-TEFb involving the 7SK small nuclear ribonucleoprotein (snRNP), factors that control both the negative and positive elongation properties of Pol II, and the mRNA processing events that are coupled with elongation are discussed.
Figures





References
-
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. - PubMed
-
- Seila AC, Core LJ, Lis JT, Sharp PA. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8:2557–2564. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

