Ubiquitin and proteasomes in transcription
- PMID: 22404630
- PMCID: PMC3637986
- DOI: 10.1146/annurev-biochem-052110-120012
Ubiquitin and proteasomes in transcription
Abstract
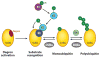
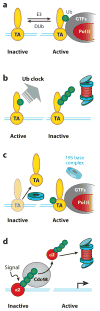
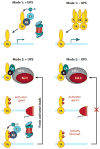
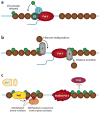
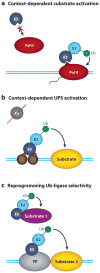
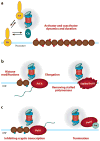
Regulation of gene transcription is vitally important for the maintenance of normal cellular homeostasis. Failure to correctly regulate gene expression, or to deal with problems that arise during the transcription process, can lead to cellular catastrophe and disease. One of the ways cells cope with the challenges of transcription is by making extensive use of the proteolytic and nonproteolytic activities of the ubiquitin-proteasome system (UPS). Here, we review recent evidence showing deep mechanistic connections between the transcription and ubiquitin-proteasome systems. Our goal is to leave the reader with a sense that just about every step in transcription-from transcription initiation through to export of mRNA from the nucleus-is influenced by the UPS and that all major arms of the system--from the first step in ubiquitin (Ub) conjugation through to the proteasome-are recruited into transcriptional processes to provide regulation, directionality, and deconstructive power.
Figures



References
-
- Ouyang J, Valin A, Gill G. Regulation of transcription factor activity by SUMO modification. Methods Mol Biol. 2009;497:141–52. - PubMed
-
- Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4:192–201. - PubMed
-
- Lipford JR, Deshaies RJ. Diverse roles for ubiquitin-dependent proteolysis in transcriptional activation. Nat Cell Biol. 2003;5:845–50. - PubMed
-
- Dennis AP, O’Malley BW. Rush hour at the promoter: how the ubiquitin-proteasome pathway polices the traffic flow of nuclear receptor-dependent transcription. J Steroid Biochem Mol Biol. 2005;93:139–51. - PubMed
-
- Baumann M, Pontiller J, Ernst W. Structure and basal transcription complex of RNA polymerase II core promoters in the mammalian genome: an overview. Mol Biotechnol. 2010;45:241–47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

