C4.4A is associated with tumor budding and epithelial-mesenchymal transition of colorectal cancer
- PMID: 22404718
- PMCID: PMC7685091
- DOI: 10.1111/j.1349-7006.2012.02263.x
C4.4A is associated with tumor budding and epithelial-mesenchymal transition of colorectal cancer
Abstract
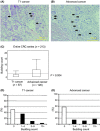
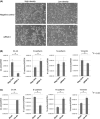
C4.4A is a glycolipid-anchored membrane protein expressed in several human malignancies. The aim of this study was to explore the association between C4.4A expression at the invasion front of colorectal cancer (CRC) and tumor budding, a putative hallmark of cell invasion of CRC. Advanced CRCs (T2-4, n = 126) had a budding count of 3.66 ± 5.66, which was significantly higher than that of T1 early CRCs (1.75 ± 2.78, n = 87). C4.4A-positive CRC specimens showed a larger budding cell number than C4.4A-negative CRC specimens in T1 CRCs, and especially advanced CRCs (9.45 ± 5.83 vs 1.60 ± 3.93). Furthermore, we found a correlation between the percentage of C4.4A-positive cases and budding count in advanced CRC. Multivariate analysis for patients' survival showed that C4.4A was superior to tumor budding as a prognostic factor. With siRNA treatment, C4.4A levels were associated with cell invasion, but not with proliferation, in HCT116 and DLD1 cell lines. An immunohistochemical study in a subset of CRCs showed no relationship between C4.4A and Ki-67 proliferation marker. In vitro assays using HCT116 indicated that C4.4A levels correlated well with epithelial-mesenchymal transition (EMT) with regard to cell morphology and alterations of EMT markers including E-cadherin, vimentin, and partially N-cadherin. We also found that C4.4A expression was significantly associated with loss of E-cadherin and gain of β-catenin in clinical CRC tissue samples. These findings suggest that a tight association between C4.4A and tumor budding may, in part, be due to C4.4A promoting EMT at the invasive front of CRC.
© 2012 Japanese Cancer Association.
Figures



Similar articles
-
Epithelial-mesenchymal transition (EMT) protein expression in a cohort of stage II colorectal cancer patients with characterized tumor budding and mismatch repair protein status.Int J Surg Pathol. 2011 Dec;19(6):751-60. doi: 10.1177/1066896911414566. Epub 2011 Jul 26. Int J Surg Pathol. 2011. PMID: 21791486
-
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis.Gut. 2013 Sep;62(9):1315-26. doi: 10.1136/gutjnl-2011-301846. Epub 2012 Jun 26. Gut. 2013. PMID: 22735571 Free PMC article.
-
N-cadherin, a novel prognostic biomarker, drives malignant progression of colorectal cancer.Mol Med Rep. 2015 Aug;12(2):2999-3006. doi: 10.3892/mmr.2015.3687. Epub 2015 Apr 27. Mol Med Rep. 2015. PMID: 25936636
-
From Dukes-MAC Staging System to Molecular Classification: Evolving Concepts in Colorectal Cancer.Int J Mol Sci. 2022 Aug 21;23(16):9455. doi: 10.3390/ijms23169455. Int J Mol Sci. 2022. PMID: 36012726 Free PMC article. Review.
-
Tumor budding in colorectal carcinoma: time to take notice.Mod Pathol. 2012 Oct;25(10):1315-25. doi: 10.1038/modpathol.2012.94. Epub 2012 Jul 13. Mod Pathol. 2012. PMID: 22790014 Review.
Cited by
-
Carbohydrate-containing molecules as potential biomarkers in colon cancer.OMICS. 2014 Apr;18(4):231-41. doi: 10.1089/omi.2013.0128. Epub 2014 Feb 6. OMICS. 2014. PMID: 24502776 Free PMC article.
-
De novo synthesis of C4.4A in hepatocellular carcinoma promotes migration and invasion of tumor cells.Oncol Rep. 2017 Nov;38(5):2697-2704. doi: 10.3892/or.2017.5980. Epub 2017 Sep 20. Oncol Rep. 2017. PMID: 29048672 Free PMC article.
-
Combined evaluation of hexokinase 2 and phosphorylated pyruvate dehydrogenase-E1α in invasive front lesions of colorectal tumors predicts cancer metabolism and patient prognosis.Cancer Sci. 2014 Sep;105(9):1100-8. doi: 10.1111/cas.12487. Epub 2014 Sep 16. Cancer Sci. 2014. PMID: 25060325 Free PMC article.
-
Characterization and function of human Ly-6/uPAR molecules.BMB Rep. 2012 Nov;45(11):595-603. doi: 10.5483/bmbrep.2012.45.11.210. BMB Rep. 2012. PMID: 23186997 Free PMC article. Review.
-
Organization, evolution and functions of the human and mouse Ly6/uPAR family genes.Hum Genomics. 2016 Apr 21;10:10. doi: 10.1186/s40246-016-0074-2. Hum Genomics. 2016. PMID: 27098205 Free PMC article. Review.
References
-
- Matzku S, Wenzel A, Liu S, Zller M. Antigenic differences between metastatic and nonmetastatic BSp73 rat tumor variants characterized by monoclonal antibodies. Cancer Res 1989; 49: 1294. - PubMed
-
- Claas C, Herrmann K, Matzku S, Mller P, Zller M. Developmentally regulated expression of metastasis‐associated antigens in the rat. Cell Growth Differ 1996; 7: 663. - PubMed
-
- Roesel M, Claas C, Seiter S, Herlevsen M, Zoeller M. Cloning and functional characterization of a new phosphatidyl‐inositol anchored molecule of a metastasizing rat pancreatic tumor. Oncogene 1998; 17: 1989–2002. - PubMed
-
- Wurfel J, Seiter S, Stassar M et al Cloning of the human homologue of the metastasis‐associated rat C4. 4A. Gene 2001; 262: 35–41. - PubMed
-
- Smith BA, Kennedy WJ, Harnden P, Selby PJ, Trejdosiewicz LK, Southgate J. Identification of genes involved in human urothelial cell‐matrix interactions: implications for the progression pathways of malignant urothelium. Cancer Res 2001; 61: 1678. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

