Antigen-presenting genes and genomic copy number variations in the Tasmanian devil MHC
- PMID: 22404855
- PMCID: PMC3414760
- DOI: 10.1186/1471-2164-13-87
Antigen-presenting genes and genomic copy number variations in the Tasmanian devil MHC
Abstract
Background: The Tasmanian devil (Sarcophilus harrisii) is currently under threat of extinction due to an unusual fatal contagious cancer called Devil Facial Tumour Disease (DFTD). DFTD is caused by a clonal tumour cell line that is transmitted between unrelated individuals as an allograft without triggering immune rejection due to low levels of Major Histocompatibility Complex (MHC) diversity in Tasmanian devils.
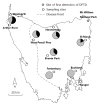
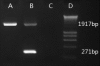
Results: Here we report the characterization of the genomic regions encompassing MHC Class I and Class II genes in the Tasmanian devil. Four genomic regions approximately 960 kb in length were assembled and annotated using BAC contigs and physically mapped to devil Chromosome 4q. 34 genes and pseudogenes were identified, including five Class I and four Class II loci. Interestingly, when two haplotypes from two individuals were compared, three genomic copy number variants with sizes ranging from 1.6 to 17 kb were observed within the classical Class I gene region. One deletion is particularly important as it turns a Class Ia gene into a pseudogene in one of the haplotypes. This deletion explains the previously observed variation in the Class I allelic number between individuals. The frequency of this deletion is highest in the northwestern devil population and lowest in southeastern areas.
Conclusions: The third sequenced marsupial MHC provides insights into the evolution of this dynamic genomic region among the diverse marsupial species. The two sequenced devil MHC haplotypes revealed three copy number variations that are likely to significantly affect immune response and suggest that future work should focus on the role of copy number variations in disease susceptibility in this species.
Figures


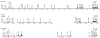





References
-
- Jones ME. In: The Encyclopedia of Mammals. Macdonald DW, editor. Oxford, UK: Oxford University Press; 2001. Large marsupial carnivores; pp. 814–817.
-
- Brown OJF. Tasmanian devil (Sarcophilus harrisi) extinction on the Australian mainland in the mid-Holocene: multicausality and ENSO intensification. Alcheringa: An Australasian Journal of Palaeontology. 2006;30:49–57. doi: 10.1080/03115510609506855. - DOI
-
- Hawkins CE, Baars C, Hesterman H, Hocking GJ, Jones ME, Lazenby B, Mann D, Mooney N, Pemberton D, Pyecroft S. et al. Emerging disease and population decline of an island endemic, the Tasmanian devil Sarcophilus harrisi. Biological Conservation. 2006;131:307–324. doi: 10.1016/j.biocon.2006.04.010. - DOI
-
- Hamede R, McCallum H, Jones ME. Seasonal, demographic and density-related patterns of contact between Tasmanian devils: Implications for transmission of Devil Facial Tumour Disease. Austral Ecology. 2008;33:614–622. doi: 10.1111/j.1442-9993.2007.01827.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

