Thermodynamic and kinetic analysis of an RNA kissing interaction and its resolution into an extended duplex
- PMID: 22404932
- PMCID: PMC3296049
- DOI: 10.1016/j.bpj.2011.12.052
Thermodynamic and kinetic analysis of an RNA kissing interaction and its resolution into an extended duplex
Abstract
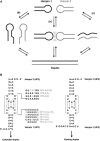
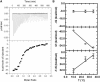
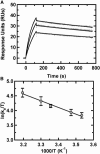
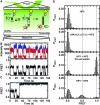
Kissing hairpin interactions form when the loop residues of two hairpins have Watson-Crick complementarity. In a unimolecular context, kissing interactions are important for tertiary folding and pseudoknot formation, whereas in a bimolecular context, they provide a basis for molecular recognition. In some cases, kissing complexes can be a prelude to strand displacement reactions where the two hairpins resolve to form a stable extended intermolecular duplex. The kinetics and thermodynamics of kissing-complex formation and their subsequent strand-displacement reactions are poorly understood. Here, biophysical techniques including isothermal titration calorimetry, surface plasmon resonance, and single-molecule fluorescence have been employed to probe the factors that govern the stability of kissing complexes and their subsequent structural rearrangements. We show that the general understanding of RNA duplex formation can be extended to kissing complexes but that kissing complexes display an unusual level of stability relative to simple duplexes of the same sequence. These interactions form and break many times at room temperature before becoming committed to a slow, irreversible forward transition to the strand-displaced form. Furthermore, using smFRET we show that the primary difference between stable and labile kissing complexes is based almost completely on their off rates. Both stable and labile complexes form at the same rate within error, but less stable species dissociate rapidly, allowing us to understand how these complexes can help generate specificity along a folding pathway or during a gene regulation event.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Mattick J.S., Makunin I.V. Non-coding RNA. Hum. Mol. Genet. 2006;15(Spec No 1):R17–R29. - PubMed
-
- Storz G., Haas D. A guide to small RNAs in microorganisms. Curr. Opin. Microbiol. 2007;10:93–95.
-
- Rana T.M. Illuminating the silence: understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:23–36. - PubMed
-
- Mello C.C., Conte D., Jr. Revealing the world of RNA interference. Nature. 2004;431:338–342. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

