Hidden multiple bond effects in dynamic force spectroscopy
- PMID: 22404941
- PMCID: PMC3296052
- DOI: 10.1016/j.bpj.2012.01.037
Hidden multiple bond effects in dynamic force spectroscopy
Abstract
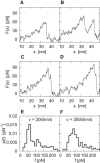
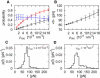
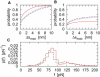
In dynamic force spectroscopy, a (bio-)molecular complex is subjected to a steadily increasing force until the chemical bond breaks. Repeating the same experiment many times results in a broad distribution of rupture forces, whose quantitative interpretation represents a formidable theoretical challenge. In this study we address the situation that more than a single molecular bond is involved in one experimental run, giving rise to multiple rupture events that are even more difficult to analyze and thus are usually eliminated as far as possible from the further evaluation of the experimental data. We develop and numerically solve a detailed model of a complete dynamic force spectroscopy experiment including a possible clustering of molecules on the substrate surface, the formation of bonds, their dissociation under load, and the postprocessing of the force extension curves. We show that the data, remaining after elimination of obvious multiple rupture events, may still contain a considerable number of hidden multiple bonds, which are experimentally indistinguishable from true single bonds, but which have considerable effects on the resulting rupture force statistics and its consistent theoretical interpretation.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Hinterdorfer P., Dufrêne Y.F. Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods. 2006;3:347–355. - PubMed
-
- Merkel R. Force spectroscopy on single passive biomolecules and single biomolecular bonds. Phys. Rep. 2001;346:343–385.
-
- Ritort F. Single-molecule experiments in biological physics: methods and applications. J. Phys. Condens. Matter. 2006;18:R531–R583. - PubMed
-
- Florin E.-L., Moy V.T., Gaub H.E. Adhesion forces between individual ligand-receptor pairs. Science. 1994;264:415–417. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

