The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain
- PMID: 22405001
- PMCID: PMC3314968
- DOI: 10.1016/j.str.2011.12.013
The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain
Abstract
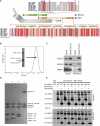
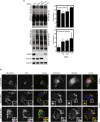
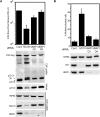
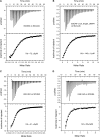
The endosomal sorting complexes required for transport (ESCRTs) facilitate endosomal sorting of ubiquitinated cargo, MVB biogenesis, late stages of cytokinesis, and retroviral budding. Here we show that ubiquitin associated protein 1 (UBAP1), a subunit of human ESCRT-I, coassembles in a stable 1:1:1:1 complex with Vps23/TSG101, VPS28, and VPS37. The X-ray crystal structure of the C-terminal region of UBAP1 reveals a domain that we describe as a solenoid of overlapping UBAs (SOUBA). NMR analysis shows that each of the three rigidly arranged overlapping UBAs making up the SOUBA interact with ubiquitin. We demonstrate that UBAP1-containing ESCRT-I is essential for degradation of antiviral cell-surface proteins, such as tetherin (BST-2/CD317), by viral countermeasures, namely, the HIV-1 accessory protein Vpu and the Kaposi sarcoma-associated herpesvirus (KSHV) ubiquitin ligase K5.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures





Comment in
-
UBAP1: a new ESCRT member joins the cl_Ub.Structure. 2012 Mar 7;20(3):383-5. doi: 10.1016/j.str.2012.02.004. Structure. 2012. PMID: 22404994 Free PMC article.
References
-
- Baumgärtel V., Ivanchenko S., Dupont A., Sergeev M., Wiseman P.W., Kräusslich H.G., Bräuchle C., Müller B., Lamb D.C. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat. Cell Biol. 2011;13:469–474. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

