Structure of the Cmr2 subunit of the CRISPR-Cas RNA silencing complex
- PMID: 22405013
- PMCID: PMC4384425
- DOI: 10.1016/j.str.2012.01.018
Structure of the Cmr2 subunit of the CRISPR-Cas RNA silencing complex
Abstract
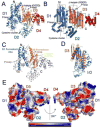
Cmr2 is the largest and an essential subunit of a CRISPR RNA-Cas protein complex (the Cmr complex) that cleaves foreign RNA to protect prokaryotes from invading genetic elements. Cmr2 is thought to be the catalytic subunit of the effector complex because of its N-terminal HD nuclease domain. Here, however, we report that the HD domain of Cmr2 is not required for cleavage by the complex in vitro. The 2.3Å crystal structure of Pyrococcus furiosus Cmr2 (lacking the HD domain) reveals two adenylyl cyclase-like and two α-helical domains. The adenylyl cyclase-like domains are arranged as in homodimeric adenylyl cyclases and bind ADP and divalent metals. However, mutagenesis studies show that the metal- and ADP-coordinating residues of Cmr2 are also not critical for cleavage by the complex. Our findings suggest that another component provides the catalytic function and that the essential role by Cmr2 does not require the identified ADP- or metal-binding or HD domains in vitro.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures




Comment in
-
Cas protein Cmr2 full of surprises.Structure. 2012 Mar 7;20(3):389-90. doi: 10.1016/j.str.2012.02.007. Structure. 2012. PMID: 22404997
Similar articles
-
Crystal structure of Cmr2 suggests a nucleotide cyclase-related enzyme in type III CRISPR-Cas systems.FEBS Lett. 2012 Mar 23;586(6):939-45. doi: 10.1016/j.febslet.2012.02.036. Epub 2012 Feb 28. FEBS Lett. 2012. PMID: 22449983
-
Crystal structure of the Cmr2-Cmr3 subcomplex in the CRISPR-Cas RNA silencing effector complex.J Mol Biol. 2013 Oct 23;425(20):3811-23. doi: 10.1016/j.jmb.2013.03.042. Epub 2013 Apr 10. J Mol Biol. 2013. PMID: 23583914
-
Structure of the Cmr2-Cmr3 subcomplex of the Cmr RNA silencing complex.Structure. 2013 Mar 5;21(3):376-84. doi: 10.1016/j.str.2013.01.002. Epub 2013 Feb 7. Structure. 2013. PMID: 23395183 Free PMC article.
-
The adenylyl and guanylyl cyclase superfamily.Curr Opin Struct Biol. 1998 Dec;8(6):770-7. doi: 10.1016/s0959-440x(98)80097-3. Curr Opin Struct Biol. 1998. PMID: 9914257 Review.
-
Structures, mechanism, regulation and evolution of class III nucleotidyl cyclases.Rev Physiol Biochem Pharmacol. 2006;157:105-40. doi: 10.1007/112_0603. Rev Physiol Biochem Pharmacol. 2006. PMID: 17236651 Review.
Cited by
-
Diversity of CRISPR systems in the euryarchaeal Pyrococcales.RNA Biol. 2013 May;10(5):659-70. doi: 10.4161/rna.23927. Epub 2013 Feb 19. RNA Biol. 2013. PMID: 23422322 Free PMC article. Review.
-
Regulation of the RNA and DNA nuclease activities required for Pyrococcus furiosus Type III-B CRISPR-Cas immunity.Nucleic Acids Res. 2020 May 7;48(8):4418-4434. doi: 10.1093/nar/gkaa176. Nucleic Acids Res. 2020. PMID: 32198888 Free PMC article.
-
Exploiting DNA Endonucleases to Advance Mechanisms of DNA Repair.Biology (Basel). 2021 Jun 14;10(6):530. doi: 10.3390/biology10060530. Biology (Basel). 2021. PMID: 34198612 Free PMC article. Review.
-
A Conserved Structural Chassis for Mounting Versatile CRISPR RNA-Guided Immune Responses.Mol Cell. 2015 Jun 4;58(5):722-8. doi: 10.1016/j.molcel.2015.05.023. Epub 2015 May 28. Mol Cell. 2015. PMID: 26028539 Free PMC article. Review.
-
HIV infection detection using CRISPR/Cas systems: Present and future prospects.Comput Struct Biotechnol J. 2023 Sep 7;21:4409-4423. doi: 10.1016/j.csbj.2023.09.005. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37711183 Free PMC article. Review.
References
-
- Artymiuk PJ, Poirrette AR, Rice DW, Willett P. A polymerase I palm in adenylyl cyclase? Nature. 1997;388:33–34. - PubMed
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

