Active human retrotransposons: variation and disease
- PMID: 22406018
- PMCID: PMC3376660
- DOI: 10.1016/j.gde.2012.02.006
Active human retrotransposons: variation and disease
Abstract
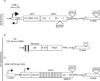
Mobile DNAs, also known as transposons or 'jumping genes', are widespread in nature and comprise an estimated 45% of the human genome. Transposons are divided into two general classes based on their transposition intermediate (DNA or RNA). Only one subclass, the non-LTR retrotransposons, which includes the Long INterspersed Element-1 (LINE-1 or L1), is currently active in humans as indicated by 96 disease-causing insertions. The autonomous LINE-1 is capable of retrotransposing not only a copy of its own RNA in cis but also other RNAs (Alu, SINE-VNTR-Alu (SVA), U6) in trans to new genomic locations through an element encoded reverse transcriptase. L1 can also retrotranspose cellular mRNAs, resulting in processed pseudogene formation. Here, we highlight recent reports that update our understanding of human L1 retrotransposition and their role in disease. Finally we discuss studies that provide insights into the past and current activity of these retrotransposons, and shed light on not just when, but where, retrotransposition occurs and its part in genetic variation.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
References
-
- Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709–732. - PubMed
-
- Romanish MT, Cohen CJ, Mager DL. Potential mechanisms of endogenous retroviral-mediated genomic instability in human cancer. Semin Cancer Biol. 2010;20:246–253. - PubMed
-
- Mayer J, Meese E. Human endogenous retroviruses in the primate lineage and their influence on host genomes. Cytogenet Genome Res. 2005;110:448–456. - PubMed
-
- Lander E, Linton L, Birren B, Nusbaum C, Zody M, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

