HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly
- PMID: 22406139
- PMCID: PMC3353549
- DOI: 10.1016/j.devcel.2012.02.001
HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly
Abstract
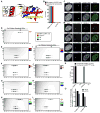
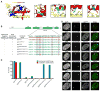
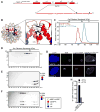
Centromeres are defined by the presence of chromatin containing the histone H3 variant, CENP-A, whose assembly into nucleosomes requires the chromatin assembly factor HJURP. We find that whereas surface-exposed residues in the CENP-A targeting domain (CATD) are the primary sequence determinants for HJURP recognition, buried CATD residues that generate rigidity with H4 are also required for efficient incorporation into centromeres. HJURP contact points adjacent to the CATD on the CENP-A surface are not used for binding specificity but rather to transmit stability broadly throughout the histone fold domains of both CENP-A and H4. Furthermore, an intact CENP-A/CENP-A interface is a requirement for stable chromatin incorporation immediately upon HJURP-mediated assembly. These data offer insight into the mechanism by which HJURP discriminates CENP-A from bulk histone complexes and chaperones CENP-A/H4 for a substantial portion of the cell cycle prior to mediating chromatin assembly at the centromere.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




References
-
- Belmont AS. Visualizing chromosome dynamics with GFP. Trends Cell Biol. 2001;11:250–257. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

