Drosophila melanogaster linker histone dH1 is required for transposon silencing and to preserve genome integrity
- PMID: 22406835
- PMCID: PMC3384340
- DOI: 10.1093/nar/gks224
Drosophila melanogaster linker histone dH1 is required for transposon silencing and to preserve genome integrity
Abstract
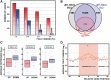
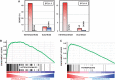
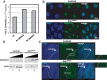
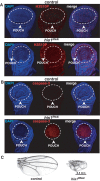
Histone H1 is an intrinsic component of chromatin, whose important contribution to chromatin structure is well-established in vitro. Little is known, however, about its functional roles in vivo. Here, we have addressed this question in Drosophila, a model system offering many advantages since it contains a single dH1 variant. For this purpose, RNAi was used to efficiently deplete dH1 in flies. Expression-profiling shows that dH1 depletion affects expression of a relatively small number of genes in a regional manner. Furthermore, depletion up-regulates inactive genes, preferentially those located in heterochromatin, while active euchromatic genes are down-regulated, suggesting that the contribution of dH1 to transcription regulation is mainly structural, organizing chromatin for proper gene-expression regulation. Up-regulated genes are remarkably enriched in transposons. In particular, R1/R2 retrotransposons, which specifically integrate in the rDNA locus, are strongly up-regulated. Actually, depletion increases expression of transposon-inserted rDNA copies, resulting in synthesis of aberrant rRNAs and enlarged nucleolus. Concomitantly, dH1-depleted cells accumulate extra-chromosomal rDNA, show increased γH2Av content, stop proliferation and activate apoptosis, indicating that depletion causes genome instability and affects proliferation. Finally, the contributions to maintenance of genome integrity and cell proliferation appear conserved in human hH1s, as their expression rescues proliferation of dH1-depleted cells.
Figures


References
-
- Bassett A, Cooper S, Wu C, Travers A. The folding and unfolding of eukaryotic chromatin. Curr. Opin. Genet. Dev. 2009;19:159–165. - PubMed
-
- Kasinky HE, Lewis JD, Dacks JB, Ausio J. Origin of H1 linker histones. FASEB J. 2001;15:34–42. - PubMed
-
- Ramakrishnan V. Histone H1 and chromatin higher-order structure. Crit. Rev. Eukaryot. Gene Expr. 1997;7:215–230. - PubMed
-
- Ramakrishnan V. Histone structure and the organization of the nucleosome. Annu. Rev. Biophys. Biomol. Struct. 1997;26:83–112. - PubMed
-
- Robinson PJJ, Rhodes D. Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr. Opin. Struct. Biol. 2006;16:336–343. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

