Codanin-1, mutated in the anaemic disease CDAI, regulates Asf1 function in S-phase histone supply
- PMID: 22407294
- PMCID: PMC3343338
- DOI: 10.1038/emboj.2012.55
Codanin-1, mutated in the anaemic disease CDAI, regulates Asf1 function in S-phase histone supply
Erratum in
- EMBO J. 2012 Jul 18;31(14):3229
Abstract
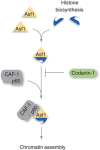
Efficient supply of new histones during DNA replication is critical to restore chromatin organization and maintain genome function. The histone chaperone anti-silencing function 1 (Asf1) serves a key function in providing H3.1-H4 to CAF-1 for replication-coupled nucleosome assembly. We identify Codanin-1 as a novel interaction partner of Asf1 regulating S-phase histone supply. Mutations in Codanin-1 can cause congenital dyserythropoietic anaemia type I (CDAI), characterized by chromatin abnormalities in bone marrow erythroblasts. Codanin-1 is part of a cytosolic Asf1-H3.1-H4-Importin-4 complex and binds directly to Asf1 via a conserved B-domain, implying a mutually exclusive interaction with the chaperones CAF-1 and HIRA. Codanin-1 depletion accelerates the rate of DNA replication and increases the level of chromatin-bound Asf1, suggesting that Codanin-1 guards a limiting step in chromatin replication. Consistently, ectopic Codanin-1 expression arrests S-phase progression by sequestering Asf1 in the cytoplasm, blocking histone delivery. We propose that Codanin-1 acts as a negative regulator of Asf1 function in chromatin assembly. This function is compromised by two CDAI mutations that impair complex formation with Asf1, providing insight into the molecular basis for CDAI disease.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


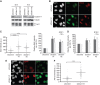
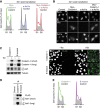
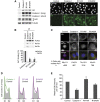
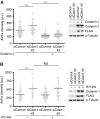
References
-
- Annunziato AT (2011) Assembling chromatin: the long and winding road. Biochim Biophys Acta 1819: 196–210 - PubMed
-
- Choudhary C, Olsen JV, Brandts C, Cox J, Reddy PN, Böhmer FD, Gerke V, Schmidt-Arras DE, Berdel WE, Müller-Tidow C, Mann M, Serve H (2009) Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol Cell 36: 326–339 - PubMed
-
- Cook AJ, Gurard-Levin ZA, Vassias I, Almouzni G (2011) A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol Cell 44: 918–927 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

