Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson's disease
- PMID: 22407749
- PMCID: PMC3403296
- DOI: 10.1002/emmm.201200215
Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson's disease
Abstract
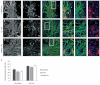
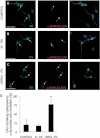
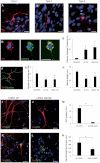
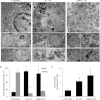
Induced pluripotent stem cells (iPSC) offer an unprecedented opportunity to model human disease in relevant cell types, but it is unclear whether they could successfully model age-related diseases such as Parkinson's disease (PD). Here, we generated iPSC lines from seven patients with idiopathic PD (ID-PD), four patients with familial PD associated to the G2019S mutation in the Leucine-Rich Repeat Kinase 2 (LRRK2) gene (LRRK2-PD) and four age- and sex-matched healthy individuals (Ctrl). Over long-time culture, dopaminergic neurons (DAn) differentiated from either ID-PD- or LRRK2-PD-iPSC showed morphological alterations, including reduced numbers of neurites and neurite arborization, as well as accumulation of autophagic vacuoles, which were not evident in DAn differentiated from Ctrl-iPSC. Further induction of autophagy and/or inhibition of lysosomal proteolysis greatly exacerbated the DAn morphological alterations, indicating autophagic compromise in DAn from ID-PD- and LRRK2-PD-iPSC, which we demonstrate occurs at the level of autophagosome clearance. Our study provides an iPSC-based in vitro model that captures the patients' genetic complexity and allows investigation of the pathogenesis of both sporadic and familial PD cases in a disease-relevant cell type.
Copyright © 2012 EMBO Molecular Medicine.
Figures



References
-
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilic J, Pekarik V, Tiscornia G, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–1284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

