Genetic control of seed shattering in rice by the APETALA2 transcription factor shattering abortion1
- PMID: 22408071
- PMCID: PMC3336138
- DOI: 10.1105/tpc.111.094383
Genetic control of seed shattering in rice by the APETALA2 transcription factor shattering abortion1
Abstract
Seed shattering is an important agricultural trait in crop domestication. SH4 (for grain shattering quantitative trait locus on chromosome 4) and qSH1 (for quantitative trait locus of seed shattering on chromosome 1) genes have been identified as required for reduced seed shattering during rice (Oryza sativa) domestication. However, the regulatory pathways of seed shattering in rice remain unknown. Here, we identified a seed shattering abortion1 (shat1) mutant in a wild rice introgression line. The SHAT1 gene, which encodes an APETALA2 transcription factor, is required for seed shattering through specifying abscission zone (AZ) development in rice. Genetic analyses revealed that the expression of SHAT1 in AZ was positively regulated by the trihelix transcription factor SH4. We also identified a frameshift mutant of SH4 that completely eliminated AZs and showed nonshattering. Our results suggest a genetic model in which the persistent and concentrated expression of active SHAT1 and SH4 in the AZ during early spikelet developmental stages is required for conferring AZ identification. qSH1 functioned downstream of SHAT1 and SH4, through maintaining SHAT1 and SH4 expression in AZ, thus promoting AZ differentiation.
Figures


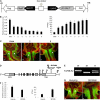


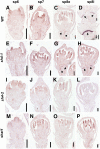

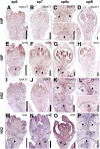
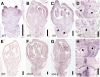
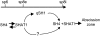
Comment in
-
SHAT1, A new player in seed shattering of rice.Plant Cell. 2012 Mar;24(3):839. doi: 10.1105/tpc.112.240310. Epub 2012 Mar 9. Plant Cell. 2012. PMID: 22408075 Free PMC article. No abstract available.
References
-
- Doebley J. (2006). Plant science. Unfallen grains: How ancient farmers turned weeds into crops. Science 312: 1318–1319 - PubMed
-
- Fuller D.Q., Qin L., Zheng Y.F., Zhao Z.J., Chen X., Hosoya L.A., Sun G.P. (2009). The domestication process and domestication rate in rice: spikelet bases from the Lower Yangtze. Science 323: 1607–1610 - PubMed
-
- Gan Y., Malhi S.S., Brandt S.A., McDonald C.L. (2008). Assessment of seed shattering resistance and yield loss in five oilseed crops. Can. J. Plant Sci. 88: 267–270
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

