A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs
- PMID: 22408077
- PMCID: PMC3336131
- DOI: 10.1105/tpc.111.095380
A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs
Erratum in
-
CORRECTION.Plant Cell. 2019 Jul;31(7):1665-1668. doi: 10.1105/tpc.19.00251. Epub 2019 May 13. Plant Cell. 2019. PMID: 31085581 Free PMC article. No abstract available.
Abstract
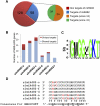
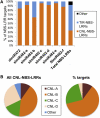
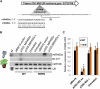
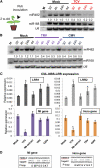
Analysis of tomato (Solanum lycopersicum) small RNA data sets revealed the presence of a regulatory cascade affecting disease resistance. The initiators of the cascade are microRNA members of an unusually diverse superfamily in which miR482 and miR2118 are prominent members. Members of this superfamily are variable in sequence and abundance in different species, but all variants target the coding sequence for the P-loop motif in the mRNA sequences for disease resistance proteins with nucleotide binding site (NBS) and leucine-rich repeat (LRR) motifs. We confirm, using transient expression in Nicotiana benthamiana, that miR482 targets mRNAs for NBS-LRR disease resistance proteins with coiled-coil domains at their N terminus. The targeting causes mRNA decay and production of secondary siRNAs in a manner that depends on RNA-dependent RNA polymerase 6. At least one of these secondary siRNAs targets other mRNAs of a defense-related protein. The miR482-mediated silencing cascade is suppressed in plants infected with viruses or bacteria so that expression of mRNAs with miR482 or secondary siRNA target sequences is increased. We propose that this process allows pathogen-inducible expression of NBS-LRR proteins and that it contributes to a novel layer of defense against pathogen attack.
Figures




Comment in
-
A microRNA cascade in plant defense.Plant Cell. 2012 Mar;24(3):840. doi: 10.1105/tpc.112.240311. Epub 2012 Mar 16. Plant Cell. 2012. PMID: 22427338 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

