Two isoforms of Geobacter sulfurreducens PilA have distinct roles in pilus biogenesis, cytochrome localization, extracellular electron transfer, and biofilm formation
- PMID: 22408162
- PMCID: PMC3347174
- DOI: 10.1128/JB.06366-11
Two isoforms of Geobacter sulfurreducens PilA have distinct roles in pilus biogenesis, cytochrome localization, extracellular electron transfer, and biofilm formation
Abstract
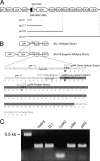
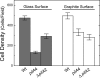
Type IV pili of Geobacter sulfurreducens are composed of PilA monomers and are essential for long-range extracellular electron transfer to insoluble Fe(III) oxides and graphite anodes. A previous analysis of pilA expression indicated that transcription was initiated at two positions, with two predicted ribosome-binding sites and translation start codons, potentially producing two PilA preprotein isoforms. The present study supports the existence of two functional translation start codons for pilA and identifies two isoforms (short and long) of the PilA preprotein. The short PilA isoform is found predominantly in an intracellular fraction. It seems to stabilize the long isoform and to influence the secretion of several outer-surface c-type cytochromes. The long PilA isoform is required for secretion of PilA to the outer cell surface, a process that requires coexpression of pilA with nine downstream genes. The long isoform was determined to be essential for biofilm formation on certain surfaces, for optimum current production in microbial fuel cells, and for growth on insoluble Fe(III) oxides.
Figures





Similar articles
-
Significance of a Posttranslational Modification of the PilA Protein of Geobacter sulfurreducens for Surface Attachment, Biofilm Formation, and Growth on Insoluble Extracellular Electron Acceptors.J Bacteriol. 2017 Mar 28;199(8):e00716-16. doi: 10.1128/JB.00716-16. Print 2017 Apr 15. J Bacteriol. 2017. PMID: 28138101 Free PMC article.
-
Aromatic amino acids required for pili conductivity and long-range extracellular electron transport in Geobacter sulfurreducens.mBio. 2013 Mar 12;4(2):e00105-13. doi: 10.1128/mBio.00105-13. mBio. 2013. PMID: 23481602 Free PMC article.
-
Anode biofilm transcriptomics reveals outer surface components essential for high density current production in Geobacter sulfurreducens fuel cells.PLoS One. 2009 May 20;4(5):e5628. doi: 10.1371/journal.pone.0005628. PLoS One. 2009. PMID: 19461962 Free PMC article.
-
Long-range electron transport to Fe(III) oxide via pili with metallic-like conductivity.Biochem Soc Trans. 2012 Dec 1;40(6):1186-90. doi: 10.1042/BST20120131. Biochem Soc Trans. 2012. PMID: 23176452 Review.
-
A long way to the electrode: how do Geobacter cells transport their electrons?Biochem Soc Trans. 2012 Dec 1;40(6):1274-9. doi: 10.1042/BST20120046. Biochem Soc Trans. 2012. PMID: 23176467 Review.
Cited by
-
Harnessing the power of microbial nanowires.Microb Biotechnol. 2018 Nov;11(6):979-994. doi: 10.1111/1751-7915.13280. Epub 2018 May 27. Microb Biotechnol. 2018. PMID: 29806247 Free PMC article. Review.
-
Synthetic Biology Toolbox, Including a Single-Plasmid CRISPR-Cas9 System to Biologically Engineer the Electrogenic, Metal-Resistant Bacterium Cupriavidus metallidurans CH34.ACS Synth Biol. 2022 Nov 18;11(11):3617-3628. doi: 10.1021/acssynbio.2c00130. Epub 2022 Oct 24. ACS Synth Biol. 2022. PMID: 36278822 Free PMC article.
-
Nitrogen Fixation and Ammonium Assimilation Pathway Expression of Geobacter sulfurreducens Changes in Response to the Anode Potential in Microbial Electrochemical Cells.Appl Environ Microbiol. 2023 Apr 26;89(4):e0207322. doi: 10.1128/aem.02073-22. Epub 2023 Mar 28. Appl Environ Microbiol. 2023. PMID: 36975810 Free PMC article.
-
Genetic analysis of electroactive biofilms.Int Microbiol. 2021 Nov;24(4):631-648. doi: 10.1007/s10123-021-00176-y. Epub 2021 Apr 27. Int Microbiol. 2021. PMID: 33907940
-
High-resolution structure of a type IV pilin from the metal-reducing bacterium Shewanella oneidensis.BMC Struct Biol. 2015 Feb 27;15:4. doi: 10.1186/s12900-015-0031-7. BMC Struct Biol. 2015. PMID: 25886849 Free PMC article.
References
-
- Alm RA, Mattick JS. 1997. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene 192:89–98 - PubMed
-
- Black WP, Xu Q, Yang Z. 2006. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol. Microbiol. 61:447–456 - PubMed
-
- Bond DR, Holmes DE, Tender LM, Lovley DR. 2002. Electrode-reducing microorganisms that harvest energy from marine sediments. Science 295:483–485 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

