The automatic annotation of bacterial genomes
- PMID: 22408191
- PMCID: PMC3548604
- DOI: 10.1093/bib/bbs007
The automatic annotation of bacterial genomes
Abstract
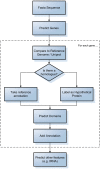
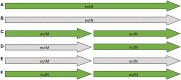
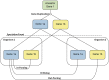
With the development of ultra-high-throughput technologies, the cost of sequencing bacterial genomes has been vastly reduced. As more genomes are sequenced, less time can be spent manually annotating those genomes, resulting in an increased reliance on automatic annotation pipelines. However, automatic pipelines can produce inaccurate genome annotation and their results often require manual curation. Here, we discuss the automatic and manual annotation of bacterial genomes, identify common problems introduced by the current genome annotation process and suggests potential solutions.
Figures

References
-
- MacLean D, Jones JD, Studholme DJ. Application of ‘next-generation' sequencing technologies to microbial genetics. Nat Rev Microbiol. 2009;7:287–96. - PubMed
-
- Stothard P, Wishart DS. Automated bacterial genome analysis and annotation. Curr Opin Microbiol. 2006;9:505–10. - PubMed
-
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. - PubMed
-
- Suzek BE, Ermolaeva MD, Schreiber M, et al. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics. 2001;17:1123–30. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

