Two programmed cell death systems in Escherichia coli: an apoptotic-like death is inhibited by the mazEF-mediated death pathway
- PMID: 22412352
- PMCID: PMC3295820
- DOI: 10.1371/journal.pbio.1001281
Two programmed cell death systems in Escherichia coli: an apoptotic-like death is inhibited by the mazEF-mediated death pathway
Abstract
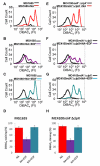
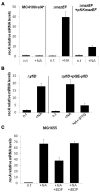
In eukaryotes, the classical form of programmed cell death (PCD) is apoptosis, which has as its specific characteristics DNA fragmentation and membrane depolarization. In Escherichia coli a different PCD system has been reported. It is mediated by the toxin-antitoxin system module mazEF. The E. coli mazEF module is one of the most thoroughly studied toxin-antitoxin systems. mazF encodes a stable toxin, MazF, and mazE encodes a labile antitoxin, MazE, which prevents the lethal effect of MazF. mazEF-mediated cell death is a population phenomenon requiring the quorum-sensing pentapeptide NNWNN designated Extracellular Death Factor (EDF). mazEF is triggered by several stressful conditions, including severe damage to the DNA. Here, using confocal microscopy and FACS analysis, we show that under conditions of severe DNA damage, the triggered mazEF-mediated cell death pathway leads to the inhibition of a second cell death pathway. The latter is an apoptotic-like death (ALD); ALD is mediated by recA and lexA. The mazEF-mediated pathway reduces recA mRNA levels. Based on these results, we offer a molecular model for the maintenance of an altruistic characteristic in cell populations. In our model, the ALD pathway is inhibited by the altruistic EDF-mazEF-mediated death pathway.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Comment in
-
In E. coli, interrupting one death pathway leads you down another.PLoS Biol. 2012;10(3):e1001278. doi: 10.1371/journal.pbio.1001278. Epub 2012 Mar 6. PLoS Biol. 2012. PMID: 22412350 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

