A genome-wide scan of Ashkenazi Jewish Crohn's disease suggests novel susceptibility loci
- PMID: 22412388
- PMCID: PMC3297573
- DOI: 10.1371/journal.pgen.1002559
A genome-wide scan of Ashkenazi Jewish Crohn's disease suggests novel susceptibility loci
Abstract
Crohn's disease (CD) is a complex disorder resulting from the interaction of intestinal microbiota with the host immune system in genetically susceptible individuals. The largest meta-analysis of genome-wide association to date identified 71 CD-susceptibility loci in individuals of European ancestry. An important epidemiological feature of CD is that it is 2-4 times more prevalent among individuals of Ashkenazi Jewish (AJ) descent compared to non-Jewish Europeans (NJ). To explore genetic variation associated with CD in AJs, we conducted a genome-wide association study (GWAS) by combining raw genotype data across 10 AJ cohorts consisting of 907 cases and 2,345 controls in the discovery stage, followed up by a replication study in 971 cases and 2,124 controls. We confirmed genome-wide significant associations of 9 known CD loci in AJs and replicated 3 additional loci with strong signal (p<5×10⁻⁶). Novel signals detected among AJs were mapped to chromosomes 5q21.1 (rs7705924, combined p = 2×10⁻⁸; combined odds ratio OR = 1.48), 2p15 (rs6545946, p = 7×10⁻⁹; OR = 1.16), 8q21.11 (rs12677663, p = 2×10⁻⁸; OR = 1.15), 10q26.3 (rs10734105, p = 3×10⁻⁸; OR = 1.27), and 11q12.1 (rs11229030, p = 8×10⁻⁹; OR = 1.15), implicating biologically plausible candidate genes, including RPL7, CPAMD8, PRG2, and PRG3. In all, the 16 replicated and newly discovered loci, in addition to the three coding NOD2 variants, accounted for 11.2% of the total genetic variance for CD risk in the AJ population. This study demonstrates the complementary value of genetic studies in the Ashkenazim.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
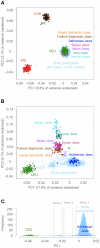
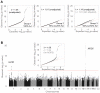

References
-
- Behar DM, Yunusbayev B, Metspalu M, Metspalu E, Rosset S, et al. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. - PubMed
-
- Price AL, Butler J, Patterson N, Capelli C, Pascali VL, et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236. doi: 10.1371/journal.pgen.0030236. - DOI - PMC - PubMed
-
- Ostrer H. A genetic profile of contemporary Jewish populations. Nat Rev Genet. 2001;2:891–898. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 DK083553/DK/NIDDK NIH HHS/United States
- NS050487/NS/NINDS NIH HHS/United States
- U01 DK062413/DK/NIDDK NIH HHS/United States
- R01 AG007992/AG/NIA NIH HHS/United States
- 2P30 DK034989/DK/NIDDK NIH HHS/United States
- R01 DK83553/DK/NIDDK NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- KL2 RR024138/RR/NCRR NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- M01 RR000425/RR/NCRR NIH HHS/United States
- P60 DK020541/DK/NIDDK NIH HHS/United States
- R01 AG018728/AG/NIA NIH HHS/United States
- M01-RR00425/RR/NCRR NIH HHS/United States
- MH080129/MH/NIMH NIH HHS/United States
- R01 DK077905/DK/NIDDK NIH HHS/United States
- NS060113/NS/NINDS NIH HHS/United States
- R01 AG7992/AG/NIA NIH HHS/United States
- R21 DK084554/DK/NIDDK NIH HHS/United States
- UL1 RR024156/RR/NCRR NIH HHS/United States
- R21 NS050487/NS/NINDS NIH HHS/United States
- U01 DK062429/DK/NIDDK NIH HHS/United States
- R01 CA137869/CA/NCI NIH HHS/United States
- R01DK077905/DK/NIDDK NIH HHS/United States
- U01 DK062422/DK/NIDDK NIH HHS/United States
- R01 NS060113/NS/NINDS NIH HHS/United States
- DK20541/DK/NIDDK NIH HHS/United States
- NS036630/NS/NINDS NIH HHS/United States
- U19 A1082713/PHS HHS/United States
- P30 DK020541/DK/NIDDK NIH HHS/United States
- P01 AG027734/AG/NIA NIH HHS/United States
- M01 RR012248/RR/NCRR NIH HHS/United States
- RC1 DK086800/DK/NIDDK NIH HHS/United States
- R01 MH080129/MH/NIMH NIH HHS/United States
- DK063491/DK/NIDDK NIH HHS/United States
- R56 NS036630/NS/NINDS NIH HHS/United States
- R01 AG-18728-01A1/AG/NIA NIH HHS/United States
- P30 DK034989/DK/NIDDK NIH HHS/United States
- R01 AG024391/AG/NIA NIH HHS/United States
- P01 DK046763/DK/NIDDK NIH HHS/United States
- U01 DK062420/DK/NIDDK NIH HHS/United States
- U01 DK062431/DK/NIDDK NIH HHS/United States
- R01 NS036630/NS/NINDS NIH HHS/United States
- DK084554/DK/NIDDK NIH HHS/United States
- M01-RR12248/RR/NCRR NIH HHS/United States
- R01 GM059507/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

