Phylo: a citizen science approach for improving multiple sequence alignment
- PMID: 22412834
- PMCID: PMC3296692
- DOI: 10.1371/journal.pone.0031362
Phylo: a citizen science approach for improving multiple sequence alignment
Abstract
Background: Comparative genomics, or the study of the relationships of genome structure and function across different species, offers a powerful tool for studying evolution, annotating genomes, and understanding the causes of various genetic disorders. However, aligning multiple sequences of DNA, an essential intermediate step for most types of analyses, is a difficult computational task. In parallel, citizen science, an approach that takes advantage of the fact that the human brain is exquisitely tuned to solving specific types of problems, is becoming increasingly popular. There, instances of hard computational problems are dispatched to a crowd of non-expert human game players and solutions are sent back to a central server.
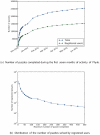
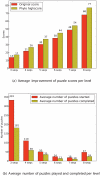
Methodology/principal findings: We introduce Phylo, a human-based computing framework applying "crowd sourcing" techniques to solve the Multiple Sequence Alignment (MSA) problem. The key idea of Phylo is to convert the MSA problem into a casual game that can be played by ordinary web users with a minimal prior knowledge of the biological context. We applied this strategy to improve the alignment of the promoters of disease-related genes from up to 44 vertebrate species. Since the launch in November 2010, we received more than 350,000 solutions submitted from more than 12,000 registered users. Our results show that solutions submitted contributed to improving the accuracy of up to 70% of the alignment blocks considered.
Conclusions/significance: We demonstrate that, combined with classical algorithms, crowd computing techniques can be successfully used to help improving the accuracy of MSA. More importantly, we show that an NP-hard computational problem can be embedded in casual game that can be easily played by people without significant scientific training. This suggests that citizen science approaches can be used to exploit the billions of "human-brain peta-flops" of computation that are spent every day playing games. Phylo is available at: http://phylo.cs.mcgill.ca.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

