Analysis of human accelerated DNA regions using archaic hominin genomes
- PMID: 22412940
- PMCID: PMC3296746
- DOI: 10.1371/journal.pone.0032877
Analysis of human accelerated DNA regions using archaic hominin genomes
Abstract
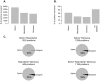
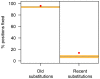
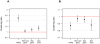
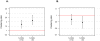
Several previous comparisons of the human genome with other primate and vertebrate genomes identified genomic regions that are highly conserved in vertebrate evolution but fast-evolving on the human lineage. These human accelerated regions (HARs) may be regions of past adaptive evolution in humans. Alternatively, they may be the result of non-adaptive processes, such as biased gene conversion. We captured and sequenced DNA from a collection of previously published HARs using DNA from an Iberian Neandertal. Combining these new data with shotgun sequence from the Neandertal and Denisova draft genomes, we determine at least one archaic hominin allele for 84% of all positions within HARs. We find that 8% of HAR substitutions are not observed in the archaic hominins and are thus recent in the sense that the derived allele had not come to fixation in the common ancestor of modern humans and archaic hominins. Further, we find that recent substitutions in HARs tend to have come to fixation faster than substitutions elsewhere in the genome and that substitutions in HARs tend to cluster in time, consistent with an episodic rather than a clock-like process underlying HAR evolution. Our catalog of sequence changes in HARs will help prioritize them for functional studies of genomic elements potentially responsible for modern human adaptations.
Conflict of interest statement
Figures

References
-
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. - PubMed
-
- Prabhakar S, Noonan JP, Paabo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

