Deep sequencing analyses of low density microbial communities: working at the boundary of accurate microbiota detection
- PMID: 22412957
- PMCID: PMC3295791
- DOI: 10.1371/journal.pone.0032942
Deep sequencing analyses of low density microbial communities: working at the boundary of accurate microbiota detection
Abstract
Introduction: Accurate analyses of microbiota composition of low-density communities (10(3)-10(4) bacteria/sample) can be challenging. Background DNA from chemicals and consumables, extraction biases as well as differences in PCR efficiency can significantly interfere with microbiota assessment. This study was aiming to establish protocols for accurate microbiota analysis at low microbial density.
Methods: To examine possible effects of bacterial density on microbiota analyses we compared microbiota profiles of serial diluted saliva and low (nares, nasopharynx) and high-density (oropharynx) upper airway communities in four healthy individuals. DNA was extracted with four different extraction methods (Epicentre Masterpure, Qiagen DNeasy, Mobio Powersoil and a phenol bead-beating protocol combined with Agowa-Mag-mini). Bacterial DNA recovery was analysed by 16S qPCR and microbiota profiles through GS-FLX-Titanium-Sequencing of 16S rRNA gene amplicons spanning the V5-V7 regions.
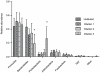
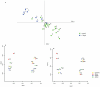
Results: Lower template concentrations significantly impacted microbiota profiling results. With higher dilutions, low abundant species were overrepresented. In samples of <10(5) bacteria per ml, e.g. DNA <1 pg/µl, microbiota profiling deviated from the original sample and other dilutions showing a significant increase in the taxa Proteobacteria and decrease in Bacteroidetes. In similar low density samples, DNA extraction method determined if DNA levels were below or above 1 pg/µl and, together with lysis preferences per method, had profound impact on microbiota analyses in both relative abundance as well as representation of species.
Conclusion: This study aimed to interpret microbiota analyses of low-density communities. Bacterial density seemed to interfere with microbiota analyses at < than 10(6) bacteria per ml or DNA <1 pg/µl. We therefore recommend this threshold for working with low density materials. This study underlines that bias reduction is crucial for adequate profiling of especially low-density bacterial communities.
Conflict of interest statement
Figures


References
-
- Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. - PubMed
-
- Salonen A, Nikkila J, Jalanka-Tuovinen J, Immonen O, Rajilic-Stojanovic M, et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods. 2010;81:127–134. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

