The evolution of filamin-a protein domain repeat perspective
- PMID: 22414427
- PMCID: PMC3728663
- DOI: 10.1016/j.jsb.2012.02.010
The evolution of filamin-a protein domain repeat perspective
Abstract
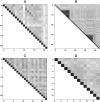
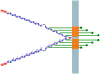
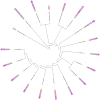
Particularly in higher eukaryotes, some protein domains are found in tandem repeats, performing broad functions often related to cellular organization. For instance, the eukaryotic protein filamin interacts with many proteins and is crucial for the cytoskeleton. The functional properties of long repeat domains are governed by the specific properties of each individual domain as well as by the repeat copy number. To provide better understanding of the evolutionary and functional history of repeating domains, we investigated the mode of evolution of the filamin domain in some detail. Among the domains that are common in long repeat proteins, sushi and spectrin domains evolve primarily through cassette tandem duplications while scavenger and immunoglobulin repeats appear to evolve through clustered tandem duplications. Additionally, immunoglobulin and filamin repeats exhibit a unique pattern where every other domain shows high sequence similarity. This pattern may be the result of tandem duplications, serve to avert aggregation between adjacent domains or it is the result of functional constraints. In filamin, our studies confirm the presence of interspersed integrin binding domains in vertebrates, while invertebrates exhibit more varied patterns, including more clustered integrin binding domains. The most notable case is leech filamin, which contains a 20 repeat expansion and exhibits unique dimerization topology. Clearly, invertebrate filamins are varied and contain examples of similar adjacent integrin-binding domains. Given that invertebrate integrin shows more similarity to the weaker filamin binder, integrin β3, it is possible that the distance between integrin-binding domains is not as crucial for invertebrate filamins as for vertebrates.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures



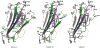



Similar articles
-
Structure of filamin A immunoglobulin-like repeat 10 from Homo sapiens.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011 Aug 1;67(Pt 8):871-6. doi: 10.1107/S1744309111024249. Epub 2011 Jul 26. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011. PMID: 21821884 Free PMC article.
-
The molecular basis of filamin binding to integrins and competition with talin.Mol Cell. 2006 Feb 3;21(3):337-47. doi: 10.1016/j.molcel.2006.01.011. Mol Cell. 2006. PMID: 16455489
-
Identification and characterization of multiple similar ligand-binding repeats in filamin: implication on filamin-mediated receptor clustering and cross-talk.J Biol Chem. 2009 Dec 11;284(50):35113-21. doi: 10.1074/jbc.M109.060954. Epub 2009 Oct 14. J Biol Chem. 2009. PMID: 19828450 Free PMC article.
-
Structural and functional aspects of filamins.Biochim Biophys Acta. 2001 Apr 23;1538(2-3):99-117. doi: 10.1016/s0167-4889(01)00072-6. Biochim Biophys Acta. 2001. PMID: 11336782 Review.
-
Filamins: promiscuous organizers of the cytoskeleton.Trends Biochem Sci. 2006 Jul;31(7):411-9. doi: 10.1016/j.tibs.2006.05.006. Epub 2006 Jun 16. Trends Biochem Sci. 2006. PMID: 16781869 Review.
Cited by
-
Small-angle X-ray scattering reveals compact domain-domain interactions in the N-terminal region of filamin C.PLoS One. 2014 Sep 22;9(9):e107457. doi: 10.1371/journal.pone.0107457. eCollection 2014. PLoS One. 2014. PMID: 25243668 Free PMC article.
-
Combining theoretical and experimental data to decipher CFTR 3D structures and functions.Cell Mol Life Sci. 2018 Oct;75(20):3829-3855. doi: 10.1007/s00018-018-2835-7. Epub 2018 May 19. Cell Mol Life Sci. 2018. PMID: 29779042 Free PMC article.
-
Identification of a Non-Pentapeptide Region Associated with Rapid Mycobacterial Evolution.PLoS One. 2016 May 5;11(5):e0154059. doi: 10.1371/journal.pone.0154059. eCollection 2016. PLoS One. 2016. PMID: 27149271 Free PMC article.
-
The tale of two talins - two isoforms to fine-tune integrin signalling.FEBS Lett. 2018 Jun;592(12):2108-2125. doi: 10.1002/1873-3468.13081. Epub 2018 May 18. FEBS Lett. 2018. PMID: 29723415 Free PMC article. Review.
-
Improved inference of tandem domain duplications.Bioinformatics. 2021 Jul 12;37(Suppl_1):i133-i141. doi: 10.1093/bioinformatics/btab329. Bioinformatics. 2021. PMID: 34252920 Free PMC article.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Andrade M, Petosa C, O'Donoghue SI, Muller CW, Bork P. Comparison of arm and heat protein repeats. J Mol Biol. 2001;309:1–18. - PubMed
-
- Andrews R, Berndt M, Lopez J. The glycoprotein Ib-IX-V complex. In: Michelson AE, editor. Platelets. Publidemic Press; 2007. pp. 145–163.
-
- Apic G, Gough J, Teichmann SA. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J Mol Biol. 2001;310:311–325. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

