Human mitotic chromosome structure: what happened to the 30-nm fibre?
- PMID: 22415369
- PMCID: PMC3321215
- DOI: 10.1038/emboj.2012.66
Human mitotic chromosome structure: what happened to the 30-nm fibre?
Abstract
EMBO J 31 7, 1644–1653 February 17 2012
The long-standing view of chromosome packaging is that 10-nm beads-on-a string chromatin fibres fold into higher-order 30-nm fibres, which further twist and coil to form highly condensed chromosomes. After four decades of intense pursuit of the structure and properties of 30-nm chromatin fibres, work of Nishino et al (2012) in this issue of The EMBO Journal demonstrates that regular 30-nm fibres are absent from human mitotic chromosomes. The emerging view is that chromosome-level condensation can be achieved through packaging of 10-nm fibres in a fractal manner.
Conflict of interest statement
The author declares that he has no conflict of interest.
Figures
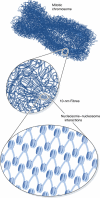
Comment on
-
Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure.EMBO J. 2012 Apr 4;31(7):1644-53. doi: 10.1038/emboj.2012.35. Epub 2012 Feb 17. EMBO J. 2012. PMID: 22343941 Free PMC article.
References
-
- Dorigo B, Schalch T, Bystricky K, Richmond TJ (2003) Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol 327: 85–96 - PubMed
-
- Fussner E, Ching RW, Bazett-Jones DP (2011) Living without 30 nm chromatin fibers. Trends Biochem Sci 36: 1–6 - PubMed
-
- Hansen JC (2002) Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms and functions. Ann Rev Biophys Biomol Str 31: 361–392 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

