Transcriptional variation in the malaria parasite Plasmodium falciparum
- PMID: 22415456
- PMCID: PMC3337437
- DOI: 10.1101/gr.129692.111
Transcriptional variation in the malaria parasite Plasmodium falciparum
Abstract
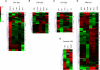
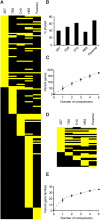
Malaria genetic variation has been extensively characterized, but the level of epigenetic plasticity remains largely unexplored. Here we provide a comprehensive characterization of transcriptional variation in the most lethal malaria parasite, Plasmodium falciparum, based on highly accurate transcriptional analysis of isogenic parasite lines grown under homogeneous conditions. This analysis revealed extensive transcriptional heterogeneity within genetically homogeneous clonal parasite populations. We show that clonally variant expression controlled at the epigenetic level is an intrinsic property of specific genes and gene families, the majority of which participate in host-parasite interactions. Intrinsic transcriptional variability is not restricted to genes involved in immune evasion, but also affects genes linked to lipid metabolism, protein folding, erythrocyte remodeling, or transcriptional regulation, among others, indicating that epigenetic variation results in both antigenic and functional variation. We observed a general association between heterochromatin marks and clonally variant expression, extending previous observations for specific genes to essentially all variantly expressed gene families. These results suggest that phenotypic variation of functionally unrelated P. falciparum gene families is mediated by a common mechanism based on reversible formation of H3K9me3-based heterochromatin. In changing environments, diversity confers fitness to a population. Our results support the idea that P. falciparum uses a bet-hedging strategy, as an alternative to directed transcriptional responses, to adapt to common fluctuations in its environment. Consistent with this idea, we found that transcriptionally different isogenic parasite lines markedly differed in their survival to heat-shock mimicking febrile episodes and adapted to periodic heat-shock with a pattern consistent with natural selection of pre-existing parasites.
Figures




Comment in
-
Adaptation: Malarial bet hedging.Nat Rev Genet. 2012 Mar 28;13(5):298-9. doi: 10.1038/nrg3224. Nat Rev Genet. 2012. PMID: 22456441 No abstract available.
References
-
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Statist Soc 57: 289–300
-
- Bethke LL, Zilversmit M, Nielsen K, Daily J, Volkman SK, Ndiaye D, Lozovsky ER, Hartl DL, Wirth DF 2006. Duplication, gene conversion, and genetic diversity in the species-specific acyl-CoA synthetase gene family of Plasmodium falciparum. Mol Biochem Parasitol 150: 10–24 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
