DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees
- PMID: 22416128
- PMCID: PMC3324026
- DOI: 10.1073/pnas.1202392109
DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees
Abstract
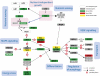
In honey bees (Apis mellifera), the development of a larva into either a queen or worker depends on differential feeding with royal jelly and involves epigenomic modifications by DNA methyltransferases. To understand the role of DNA methylation in this process we sequenced the larval methylomes in both queens and workers. We show that the number of differentially methylated genes (DMGs) in larval head is significantly increased relative to adult brain (2,399 vs. 560) with more than 80% of DMGs up-methylated in worker larvae. Several highly conserved metabolic and signaling pathways are enriched in methylated genes, underscoring the connection between dietary intake and metabolic flux. This includes genes related to juvenile hormone and insulin, two hormones shown previously to regulate caste determination. We also tie methylation data to expressional profiling and describe a distinct role for one of the DMGs encoding anaplastic lymphoma kinase (ALK), an important regulator of metabolism. We show that alk is not only differentially methylated and alternatively spliced in Apis, but also seems to be regulated by a cis-acting, anti-sense non-protein-coding transcript. The unusually complex regulation of ALK in Apis suggests that this protein could represent a previously unknown node in a process that activates downstream signaling according to a nutritional context. The correlation between methylation and alternative splicing of alk is consistent with the recently described mechanism involving RNA polymerase II pausing. Our study offers insights into diet-controlled development in Apis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Wilson EO. The Insect Societies. Cambridge, MA: Harvard Univ Press; 1971.
-
- Hartfelder K, Engels W. Social insect polymorphism: Hormonal regulation of plasticity in development and reproduction in the honeybee. Curr Top Dev Biol. 1998;40:45–77. - PubMed
-
- Evans JD, Wheeler DE. Gene expression and the evolution of insect polyphenisms. Bioessays. 2001;23:62–68. - PubMed
-
- Maleszka R. Epigenetic integration of environmental and genomic signals in honey bees: The critical interplay of nutritional, brain and reproductive networks. Epigenetics. 2008;3:188–192. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

